In this issue of Blood, Speedy et al identify germ line loss-of-function mutations involving the telomere shelterin complex in a subset of families with chronic lymphocytic leukemia (CLL).1
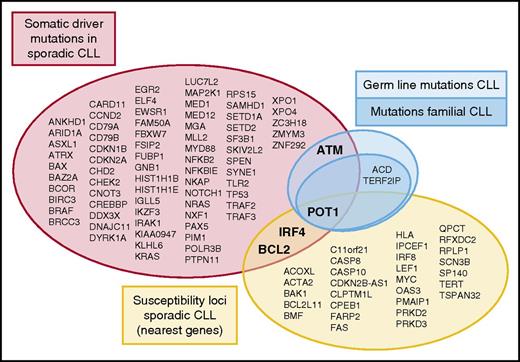
Genetic alterations involved in the pathogenesis of CLL. Different studies in large series of sporadic CLL have identified somatic driver mutations (red)7 ,8 and susceptibility loci (yellow) related to the pathogenesis of the disease. Furthermore, germ line mutations outside a familiar context in CLL have been identified in ATM (light blue). Speedy et al, for the first time, describe germ line mutations in a familial context in CLL targeting the shelterin complex (dark blue; POT1, TERF2IP, and ACD). The overlap between genetic variants at different levels (somatic driver mutations, susceptibility loci, and germ line mutations) as indicated comprises just a handful of genes.
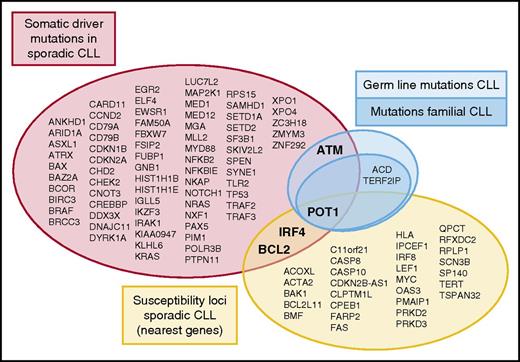
Genetic alterations involved in the pathogenesis of CLL. Different studies in large series of sporadic CLL have identified somatic driver mutations (red)7 ,8 and susceptibility loci (yellow) related to the pathogenesis of the disease. Furthermore, germ line mutations outside a familiar context in CLL have been identified in ATM (light blue). Speedy et al, for the first time, describe germ line mutations in a familial context in CLL targeting the shelterin complex (dark blue; POT1, TERF2IP, and ACD). The overlap between genetic variants at different levels (somatic driver mutations, susceptibility loci, and germ line mutations) as indicated comprises just a handful of genes.
CLL has the highest risk among lymphoid neoplasms of occurring in relatives of affected individuals.2 Susceptibility to CLL has been linked to different chromosomal loci. Although some CLL patients may carry germ line mutations in ATM,3 no genetic alterations or specific variants related to familial CLL have been discovered thus far. Speedy et al now report for the first time identification of germ line mutations in POT1 that cosegregated with CLL in relatives of 4 families. In addition, the authors found the presence of germ line mutations involving other elements of the shelterin complex in 3 other CLL families, namely in TERF2IP (2 families) and ACD (1 family). Their finding supports the importance of telomere function in the pathogenesis of CLL. Genetic alterations involving different proteins regulating this complex chromosomal structure have now been identified in CLL at 3 different levels: somatic mutations in sporadic CLL (POT1), single nucleotide polymorphisms with increased risk to develop the disease (POT1 and TERT), and now germ line mutations in families cosegregating with the disease (POT1, TERF2IP, and ACD).
The shelterin complex is a crucial regulator of telomere function, modulating telomere replication, extension by telomerase, and protecting the ends of the chromosomes from degradation or aberrant recombination.4 The structural predictions of Speedy et al suggest that the identified mutations in these genes reduce their protein:DNA and protein:protein binding affinity and therefore potentially interfere with their telomeric function. A somatic CLL mutation in the same residue Tyr36 observed in one of the families studied by Speedy et al previously was proven to disrupt POT1 binding to DNA.5 One of the open questions is how the aberrant shelterin complex contributes to CLL development. The current study shows that the telomere length was similar in cases with and without germ line shelterin mutations, as was also observed comparing sporadic CLL cases with and without somatic POT1 mutations.5 Functional studies in cell lines and observations in primary CLL cells, however, clearly show that POT1 mutated cells accumulate numerous telomeric and chromosomal abnormalities that may be responsible, at least in part, for the progression of the disease.5 Intriguingly, a recent study linked increased risk of development of CLL and other lymphoid neoplasms to longer genetically determined telomere length of peripheral blood leukocytes, emphasizing again the link between telomere function and the development of disease.6
Because the risk of CLL associated with the POT1 p.Gln376Arg mutation was increased 3.61-fold, it suggests that shelterin gene mutations have moderate penetrance. Such an assertion is supported by the observation that the unmutated allele is not lost in the CLL cells of mutated carriers. Determination of the true penetrance of these variants will likely require the study of the risk loci in unaffected individuals, for example, the siblings of the affected patients, although ethical considerations may limit this analysis.
The understanding of the role of genetic alterations in the pathogenesis of CLL has been expanded in the last few years with extensive genome wide analysis focusing on 2 major aspects. Whole genome/exome sequencing efforts have elucidated the landscape of somatic mutations in untreated CLL patients, with the identification of ∼80 highly confident driver genes (see figure).7,8 On the other hand, genome-wide association studies (GWASs) have identified 31 susceptibility loci conferring an increased risk of developing the disease (see figure and results of 8 comprehensive GWASs referred to by Speedy et al). Some of these loci are within or in the vicinity of genes whose function may influence the development of the disease, but most of them are in noncoding regions, and their direct role in the pathogenesis of CLL is not understood. Most genes identified in both types of studies are different, and only POT1, BCL2, and IRF4 are found in both subsets of genes (see figure). The small group of genes in the overlap between the 2 subsets is intriguing. Possible reasons for this minor overlap are that genes related to the respective susceptibility loci and somatic mutated genes may act at different moments in the development of the disease. On the other hand, although the genes in both subsets are different, they may be targeting similar pathways. For example, germ line variations related to TERT interfere with telomeric function, which are also targeted by somatic mutations in POT1, frequently found in CLL.
A challenge for the future will be to detect underlying germ line alterations in the other CLL families studied for which thus far no germ line mutations have been detected. Potentially, germ line mutations may be found in noncoding regions. Intriguingly, it is becoming clear that noncoding regions in the genome may affect gene expression by interaction with their target genes in 3-dimensional (3D) space in the nucleus.9 Consequently, genetic alterations within noncoding regions may affect distant target genes. This was shown for somatic mutations in a distant PAX5 enhancer that significantly reduced the expression of this master B-cell regulator in affected CLL cases.7 Similarly, the functional and 3D analysis of the CLL susceptibility locus located in an enhancer at 15q15.1 was identified to target the antiapoptotic BCL2 pathway, by influencing the expression of BMF in CLL.10 Hence, an integrative analysis of genetic screens together with gene expression and comprehensive epigenetic studies would be instrumental to better understand the role of susceptibility loci and somatic mutations in noncoding regions in the pathogenesis of CLL.
Conflict-of-interest disclosure: The authors declare no competing financial interests.