Abstract
BRCA1 is one of the most important gene associated with familial breast and ovarian cancer susceptibility and is involved in the DNA damage repair and cell cycle arrest. Alterations in BRCA1 and their consequences are well characterized in breast and ovarian tumors, while little is known about its role in Acute Myeloid Leukemia (AML).
We aimed to investigate the frequency and the interplay of BRCA1 alterations and patterns of somatic mutations (SNVs) and copy number variations (CNVs) of in AML patients.
We genotyped 118 AML samples at either diagnosis or relapse by Single Nucleotide Polymorphism (SNP) array (SNP6.0 and Cytoscan HD, Affymetrix) to detect CNVs. In addition, we performed Whole-Exome Sequencing (WES, 100 bp paired-end, Illumina) of 56 genotyped AML patients to detect SNVs and small indels (MuTect and Varscan 2.0). Differences in survival were assessed using Kaplan-Meier survival analysis and Long-Rank test, or Breslow when indicated.
We detected BRCA1 loss in 14 out of 118 patients (12%), ranging from 1,6 Kb to the loss of the entire chromosome 17, with none of the BRCA1 loss patients having a normal karyotype. Moreover, BRCA1 losses significantly co-occurred with PALB2 and RAD50 losses. Notably, the PALB2 loss (chromosome 16) mostly involved exon 12 and was detected also in patients without the BRCA1 loss, with a frequency of 11 out of 118 cases (9%), suggesting a pivotal and previously undescribed role in AML pathogenesis. BRCA1 loss patients were enriched for copy number alterations in genes involved in the regulation of mitotic recombination, DNA repair and positive regulation of kinase activity compared with BRCA1 wild-type cases (p <0.01). To investigate which genes and biological functions cooperates with the pathways affected by structural alterations, we integrated the analysis with WES data on 56 out of 118 AML patients. We focused on the genes involved in the DNA damage repair (DDR) and we identified somatic SNVs in BRCA1, TP53 and in the Fanconi Anemia genes BRCA2 and FANCE. Analysis of co-occurrence and mutual exclusivity revealed that BRCA1 loss and DDR-related alterations (BRCA1, PALB2, BRCA2, and FANCE) were significantly associated with TP53 alterations (p=0.0001 and p=0.0003, respectively). In addition, alterations in the DDR genes were mutually exclusive with mutations in KRAS, IDH1/2, BCOR, BCORL1, NPM1 and myeloid transcription factors as RUNX1 and CEBPA. Notably, patients without alteration in the DDR genes were enriched for mutations in DNMT3A and FLT3.
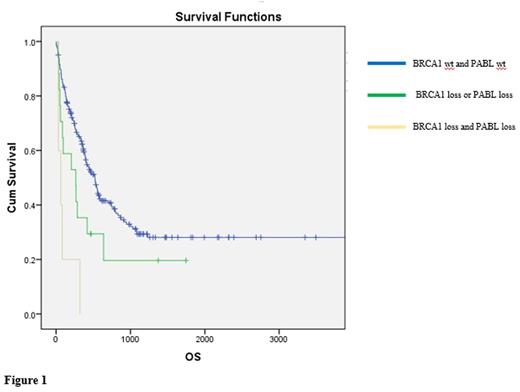
Finally, BRCA1 and PALB2 alterations defined group of patients with poor overall survival (OS). In particular, patients with alterations of either BRCA1 or PALB2 had a worst prognosis compared to patients without alterations in the two genes (p=0.009 and p=0.001, respectively). Patients harboring PALB2 loss had a poorer prognosis compared with patients with complex karyotype (p=0.021, Breslow test). In addition, double BRCA1 and PALB2 loss, single BRAC1 or PALB2 hits and lack of alterations in BRCA1 and PALB2 defined three different classes of risks in our AML cohort (p<0.001).
In summary, we define for the first time a novel subgroup of AML patients characterized by alterations targeting the DDR pathway, and in particular the BRCA1 and PALB2 genes. Our data suggest that a signature of genes involved in the DDR and cell cycle regulation synergize and led to the uncontrolled proliferation, independently of DNA damage accumulation. Our results may help improve patient stratification and define ad hoc therapeutic strategies for this aggressive leukemia type.
Acknowledgments: ELN, AIL, AIRC, progetto Regione-Università 2010-12 (L. Bolondi), Fondazione del Monte di Bologna e Ravenna, FP7 NGS-PTL project.
Guadagnuolo:CellPly S.r.l.: Employment. Martinelli:Novartis: Speakers Bureau; Genentech: Consultancy; Roche: Consultancy, Speakers Bureau; BMS: Speakers Bureau; Celgene: Consultancy, Speakers Bureau; Amgen: Consultancy, Speakers Bureau; MSD: Consultancy; Pfizer: Consultancy, Speakers Bureau; Ariad: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.