Abstract
AML is a hematological disorder resulting from proliferation and expansion of malignant myeloid cells. Clinical outcome for AML remains dismal despite intensive therapy in part due to the disease heterogeneity with various cytogenetic and molecular lesions. Fms-Like Tyrosine Kinase-3 (FLT3) is a receptor tyrosine kinase expressed hematopoietic stem/progenitor cells. Activating mutations of FLT3 gene due to internal tandem duplication of the juxtamembrane domain coding sequence (FLT3/ITD) causes autonomous cellular proliferations leading to disease progression. Metabolomic profiling has been successfully utilized to identify metabolic alterations in hematological disorders. However, no studies on metabolic alterations associated with pediatric AML have been reported at this time. In this study we propose to establish the metabolomic landscape in pediatric AML patients and identify differential expression of metabolites based on FLT3/ITD status.
Cellular and plasma metabolomics profile was generated from 32 matching diagnostic material from 16 patients with and without FLT3/ITD (N=8 for each cohort and each sample was run in duplicate) treated on COG-AAML0531 study. Global metabolomics profiling was performed on a Thermo Q-Exactive Oribtrap mass spectrometer with Dionex UHPLC and autosampler. All samples were analyzed in duplicate in positive and negative heated electrospray ionization with a mass resolution of 35,000 at m/z 200 as separate injections. Separation was achieved on an ACE 18-pfp 100 x 2.1 mm, 2 µm column with mobile phase A as 0.1% formic acid in water and mobile phase B as acetonitrile. This is a polar embedded stationary phase that provides comprehensive coverage, but does have some limitation is the coverage of very polar species. The flow rate was 350 µL/min with a column temperature of 25¡C. 4 µL was injected for negative ions and 2 µL for positive ions. Statistical analysis was performed using MetaboAnalyst software using all metabolites (known and unknown) as well as only the annotated metabolites. Univariate analysis was performed by volcano plot and Multivariate analysis was performed using PCA, PLS-DA and OPLS-DA.
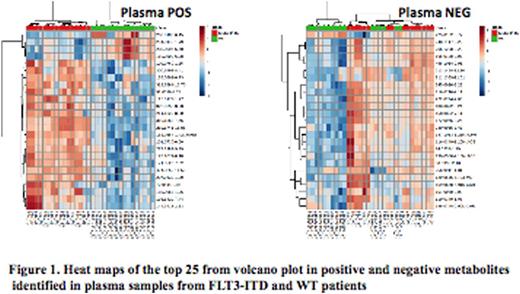
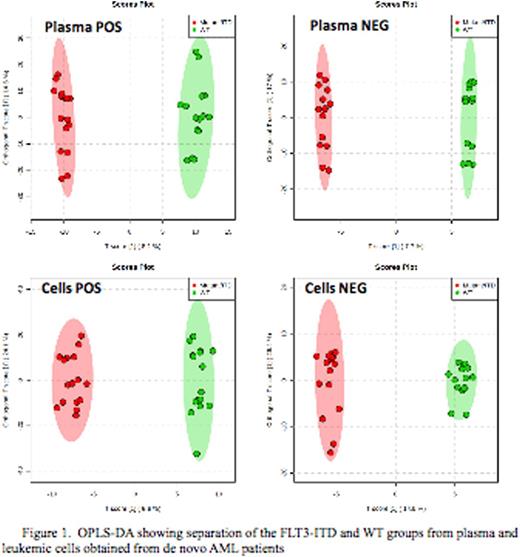
Total of 2966 plasma metabolome (779 negative and 2187 positive) and 1742 (227 negative and 1515 positive) cellular metabolome features were identified. All subsequent data analyses were normalized to the sum of metabolites for each sample. Comparison of the cellular metabolome in patients with and without FLT3/ITD identified 12 known and 135 unknown metabolites that were significantly different between two cohorts (p<0.05). Similar comparison of the plasma metabolome identified19 known and 300 unknown metabolites in the patient with and without FLT3/ITD (top results are shown in Fig.1). Orthogonal partial least squares-discriminant analysis (OPLS-DA) showed clear separation between the 2 groups (Fig.2). Some of the top known plasma metabolites (p<0.01) differentiating patients by FLT3 status include guanine, pyrimidine-2-3dicarboxylate, acetylglycine, acetyl-L-alanine, aminobutonate gaba, L-carnitine, methyl-2 oxovaleric acid, asparagine, acetyl arginine, Hydroxydecanoic acid, cysteic acid and glycocholic acid. Within leukemic cells top metabolites differentiating between FLT3 status included actyly carnitine, adenosine monophosphate, hypoxanthine, diaminohepatanedioate, guanine and sphingosine.
Metaboanalyst Pathway analysis module mapped the differentiating metabolites to aminoacyl-tRNA biosynthesis, Glycerophospholipid metabolism, Cysteine and methionine metabolism, Pantothenate and CoA biosynthesis, Purine metabolism.
This pilot study defines distinct metabolomics signature associated with genomic subtype of AML (FLT3/ITD). As metabolomics provides an insight into the ultimate metabolic destination of normal and malignant hematopoiesis, it has a potential to provide a unique insight into the altered metabolic pathways in AML and identify pathways and networks that might be shared by varied genomic subtypes of AML. Such data can help merge rare genomic variants based on shared metabolic signatures and more appropriately guide directed therapies.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.