Abstract
Introduction
Acute lymphoblastic leukaemia (ALL) in infants has poor overall survival despite being characterized by very few genetic aberrations per case. The most common genetic change, present in over 75% of cases, is the rearrangement of the mixed lineage leukaemia (MLL/KMT2A) gene (MLL-R) that also occurs in AML and mixed phenotype acute leukaemia (MPAL). Because infant ALL is rare and distinctive, most Australian and New Zealand patients are enrolled on specific clinical trials. In earlier infant ALL trials, MRD was not used for risk stratification firstly because of the difficulty of finding sensitive markers for specific immunoglobulin and T-cell receptor (Ig/TCR) gene rearrangements and the secondly because, in infant ALL, Ig/TCR markers are often present in sub-clones that can be lost at relapse due to clonal selection. MRD testing is performed in the current Interfant 06 trial preferentially using markers based on the genomic breakpoint sequences of MLL gene rearrangements. The treatment of infant ALL remains very challenging with relatively poor survival rates attributable to both toxicity and relapse. The objectives of this study were therefore to analyse MRD data for Interfant 06 patients enrolled at ANZCHOG centres and to perform a pilot experiment evaluating gene expression for key genes in infant ALL samples on a microfluidics platform, as a basis for identifying potential targeted therapies.
Methods
MRD was measured in bone marrow DNA from Interfant 06 patients enrolled since 2006, using sensitive Real-time Quantitative PCR (PCR-MRD) patient-specific assays to detect either MLL gene rearrangements (in 17) and/or conventional immunoglobulin and T-cell receptor (Ig/TCR) markers (21). Gene expression levels for 90 genes important in childhood cancers were measured by Taqman-based microfluidic assays in duplicate using cDNA from 2 micrograms of total RNA from 10 infant ALL samples (5 MLL, 5 non-MLL) at diagnosis (6) or relapse (4).
Results
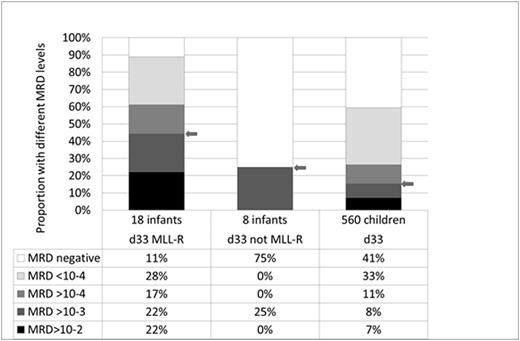
Patient-specific PCR-MRD tests were developed for 27 out of 28 Australian and New Zealand infant ALL patients. 17/18 MLL-R ALL patients had MLL-R assays and 21/28 patients had Ig/TCR MRD tests, with only 1 (non-MLL) patient having no MRD markers. There was a wide range of MRD responses to induction therapy (Figure 1). Bone marrow MRD at the end of induction was high (>1x10-3) in 44% of MLL-R ALL infants compared to 25% of non-MLL ALL infants and 15% of older children enrolled on ANZCHOG ALL8. In 8/13 MLL-R patients who had both types of marker, MRD levels were higher when measured by their MLL-R marker than by their Ig/TCR marker. In a set of 90 genes selected for expression analysis, higher levels were found for 17 genes in 2 or more of the 10 infant ALL samples evaluated. These more highly expressed genes included potential or known drug targets BCL2, ERBB2, ERBB4, ILRA2, CSF1R and PARP1.
Conclusions
The quantitation of MRD based on MLL rearrangements in ALL is effective and can also be used to monitor response to therapy in infant ALL as well as MLL-R cases of AML and MPAL. The combined application of MLL-R and Ig/TCR markers allowed 97% of infant ALL patients to be MRD monitored with a sensitive marker. In most patients with both type of MRD marker, higher levels of MRD were detected in end of induction samples using the MLL versus Ig/TCR tests. One interpretation is that Ig/TCR genes are rearranged after the MLL rearrangement in ALL sub-clones that are both more mature and more chemo-sensitive. This finding also confirms the current consensus that disease-related MLL-R markers provide better risk assessment than Ig/TCR markers. Our Fluidigm analysis has shown that quantitative measurement of multiple gene expressions is feasible on small RNA samples and can be used to rapidly screen for specific expression of genes coding for drug targets in ALL patients.
Support: NHMRC Australia APP1057746, Sporting Chance Cancer Foundation.
Comparison of MRD response to induction therapy in MLL-R infant ALL compared with non-MLL infant ALL (Interfant 06) and older children (ANZCHOG ALL8).
Comparison of MRD response to induction therapy in MLL-R infant ALL compared with non-MLL infant ALL (Interfant 06) and older children (ANZCHOG ALL8).
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.