Abstract
Background:TP53 mutations arise in a broad set of hematologic diseases and are associated with poor prognosis and therapy failure. Even small TP53 mutated clones were demonstrated to be of clinical relevance and therefore their early detection is mandatory. Mutations can occur throughout the entire gene (mainly exons 4-10) and include base exchanges, deletions and insertions. Next generation sequencing (NGS) generally detects mutations, which are present in at least 3% of sequences. The detection of mutations at burdens below 3% is still hampered by polymerase and sequencing errors. Especially true mutations caused by single base exchanges (missense-, nonsense- and splice site mutations) are difficult to distinguish from non-specific background. To overcome this limitation, individual molecules can be tagged by unique molecular identifiers (UMIs). UMIs are small random sequences added to each individual molecule before amplification. Building a consensus sequence out of all amplified products derived from one original DNA template, allows reconstructing the sequence of the initial molecule and thereby eliminates amplification errors in silico.
Aim: 1) To evaluate reduction of non-specific background in NGS assays after using UMIs to build consensus sequences. 2) To use consensus sequencing to identify early subclones by backtracking of known TP53 mutations in CLL patients.
Methods: We added eight random nucleotides as UMI to all reverse primers (design adapted from Peng, et al. 2015) and performed an initial primer extension step. In a subsequent PCR, regions of interest including UMIs were amplified and adapters were added for MiSeq sequencing (Illumina, San Diego, CA). We performed 119 sequencing assays, and analyzed results with a 1% detection limit. Consensus sequences were built using SeqNext 4.3 (JSI Medical Systems, Kippenheim, Germany). A median number of 6 (range: 2-115) reads/consensus read was obtained. Samples contained 66 previously identified mutations (47 single base exchanges; 14 small deletions; 5 small insertions), of which 29 had a burden of 3% or lower. These had been confirmed to be specific by independently repeated sequencing analysis or known in patients at multiple time points during follow-up.
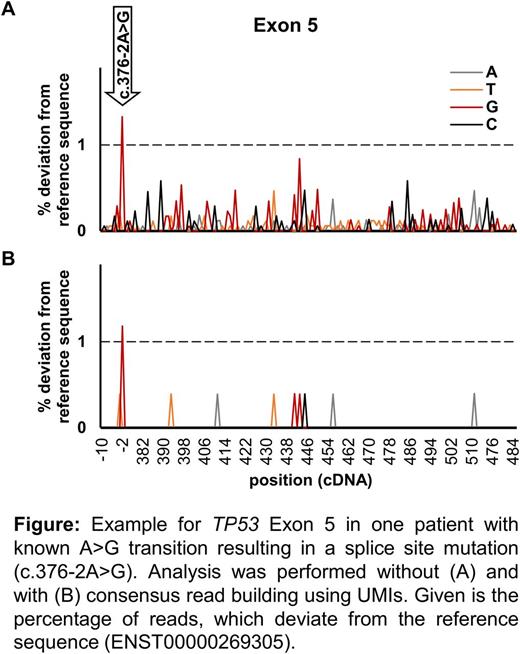
Results: We evaluated background signal at each position before consensus read building. On average 97.5% of bases had low-level non-specific background. At each individual position, 0.01-0.87% of reads deviated from the reference sequence (Figure A). Building consensus reads by combining all sequences derived from one original DNA molecule reduced the number of bases with low-level non-specific signal to an average of 18% (Figure B).
We performed 119 sequencing analysis with a 1% detection limit, aiming to identify 66 known mutations and no non-specific deviations from the reference sequence. With both approaches, 63/66 mutations were detected and correlation of mutation burdens was high (R2=0.99; P<0.001, calculated by linear regression). Two mutations (1% and 2%) only reached the detection cutoff without consensus read building and one mutation only after consensus read building. Importantly, without using UMIs for consensus reads, in sum 88 non-specific deviations from the reference sequence (likely artifacts) were detectable with the 1% cutoff, while only two likely artifacts remained after consensus sequences building.
Thus, the reduction of sequencing artifacts should allow using a 1% cutoff for mutation detection in future routine settings. Therefore, we conducted a backtracking using 31 of the above identified TP53 mutations detected in 15 CLL patients. Samples covered a median timeframe of 26 (range: 2-68) months. Using the previous 3% cutoff for mutation burden, we identified 8/31 (26%) mutations 2-55 months earlier (median: 4 months). Even more, using the adjusted 1% cutoff with UMIs allowed detecting 18/31 (58%) TP53 mutations earlier (2-55 months; median: 18 months).
Conclusion: Including UMIs and building consensus sequences, 1) reduced background signal in silico and 2) allowed improving NGS detection limits. This is crucial for the identification of low burden mutations in TP53 and other genes, where small subclones can rapidly expand and have been shown to require early treatment intervention.
Baer:MLL Munich Leukemia Laboratory: Employment. Nadarajah:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.