Abstract
Introduction: Myelodysplastic syndromes (MDS) are frequently associated with somatic mutations in epigenetic modifiers such as de novo methyltransferase 3A (DNMT3A). However, so far the significance of specific epigenetic modifications for disease stratification remains largely unknown. In this study, we investigated if epigenetic biomarkers, which were previously described to be relevant in acute myeloid leukemia (AML), are also of prognostic impact in MDS.
Methods: Peripheral blood samples of MDS patients (n=126; f/m=59/67; median age 66; range 26-93) equally distributed across all risk groups based on the revised International Prognostic Scoring System (IPSS-R; very low/low=43; int=37; high/very high=43; n.a.=3) were analyzed at initial diagnosis. Genomic DNA was isolated, bisulfite converted, and DNA methylation (DNAm) level at selected genomic regions were determined by pyrosequencing as described before: (1) hypermethylation at a CpG site in complement component 1 subcomponent R (C1R), (2) an epigenetic age prediction with an Epigenetic-Aging-Signature based on three CpG sites located in the genes ITGA2B, ASPA and PDE4C, and (3) an epimutation in the DNMT3A locus, mimicking somatic mutations of this gene, were all reported to correlate with overall survival (OS) in AML patients. Results were subsequently compared to clinical parameters such as IPSS-R, leukemic progression, and OS.
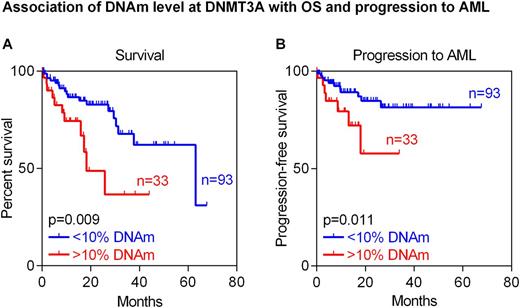
Results: A clear tendency for longer OS of MDS patients was observed if DNAm level at C1R was above median (22%; two-year survival 67% [95% CI 53-84%] in hypo- vs. 84% [95% CI 74-95%] in hypermethylated samples; P=0.071), which is in line with previous findings in AML samples. The predicted epigenetic age determined by the Epigenetic-Aging-Signature correlated moderately with the chronological age of the investigated MDS patients (R=0.42) and their OS (P=0.029). This effect was also seen in a multivariable analysis of this cohort including predicted and chronological age (P=0.040). Finally, we stratified MDS patients by the DNAm level of 10% in DNMT3A. Similar to AML, also MDS patients with higher methylation at the CpG site represented on a microarray (cg23009818) showed in tendency shorter OS (two-year survival 79% [95% CI 69-89%] in hypo- vs. 65% [95% CI 45-93%] in hypermethylated samples; P=0.110). In fact, this association was even more pronounced at a neighboring CpG site (two-year survival 83% [95% CI 74-92%] in hypo- vs. 49% [95% CI 29-84%] in hypermethylated samples; P=0.009; Figure A). Moreover, increased DNAm level at this neighboring CpG site in DNMT3A was indicative for progression into AML (after two years: 15% [95% CI 6-24%] in hypo- vs. 44% [95% CI 12-76%] in hypermethylated samples; P=0.011; Figure B). Of note, none of these markers correlated with IPSS-R categories indicating that they might provide independent prognostic parameters.
Conclusion: The analyzed epigenetic biomarkers revealed prognostic relevance in MDS patients and we suggest considering them in future risk stratification models. Particularly the aberrant hypermethylation of DNMT3A, which may also result in alternative splicing of DNMT3A transcripts, was associated with accelerated leukemic progression and shorter OS.
Božić:Cygenia GmbH: Consultancy. Wagner:Cygenia GmbH: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.