Abstract
Introduction:
Proteasome inhibition is the backbone of various Multiple Myeloma (MM) treatment regimens, leading to durable responses and high quality remissions. However, under prolonged therapy patients eventually develop drug resistance and the underlying mechanisms have been poorly understood. Proteasome inhibitor resistant cell lines were generated by continuous exposure to proteasome inhibiting drugs. In these cell line models a number of variants within the PSMB5 gene were observed. This includes single point mutations, leading to conformational changes of the _5 subunit within the 20S proteolytic core of the proteasome, impairing its chymotryptic catalytic function and the binding of inhibitory drugs. However, no PSMB5 mutations could, so far, be identified in human disease, leaving the functional relevance of such mutations to be determined.
Methods:
We applied the MM Mutation Panel (M3P) and used the Personal Genome Machine (Life Technologies) to sequence CD138+ purified bone marrow plasma cells and peripheral blood germ line control from a MM patient in third relapse that had been previously treated by various proteasome inhibitor containing therapies (VTD-PACE, VCD, PAD-Rev). This patient was subsequently treated with a pomalidomide, bortezomib, adriamycin and dexamethasone combination therapy (Pom-PAD) and achieved a partial remission after 4 cycles of therapy. When being exposed to VCD at earlier relapse a complete remission was induced within 6 cycles of therapy, demonstrating excellent response to proteasome inhibition at this earlier disease stage.
Results:
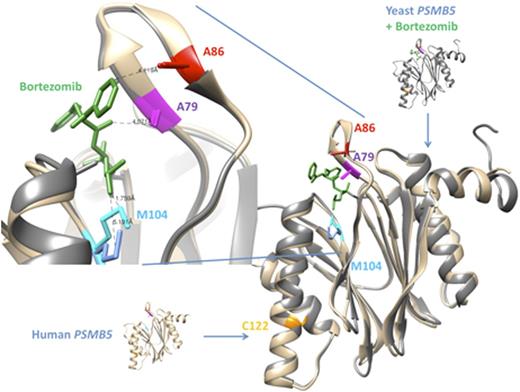
We identified clonal missense mutations in this patient at known NRAS hotspot location (p.Gln61Arg) and in MAX (p.Arg35His). Most notably we found and validated four subclonal single point mutations in PSMB5, each of them in subclonal frequencies with variant reads (VR) ranging from 1.9%- 5.9% (average read depth 750X). All PSMB5 mutations occurred in a highly conserved region in exon 2 of the gene (p.Cys122Tyr, p.Met104Ile, p.Ala86Pro, p.Ala79Thr), with three of them being located within the proteasome inhibitor drug binding site. Our 400bp amplicon design allowed us to observe that each mutation identified in PSMB5 is exclusively present on a different sequencing read and no reads are shared between the mutations. This implies that the mutations are present on different subclones of the tumor, which means that, despite low VR frequencies of the single mutation, in sum more than a quarter of the whole tumor might be affected by mutated PSMB5. At a disease stage when the patient was well responsive to proteasome inhibitor treatment (at diagnosis and at first relapse), of note, none of the mutations in PSMB5 is detectable.
Conclusion:
Here we report the first in human PSMB5 mutation in a MM patient. These mutations evolved in parallel within different subclones of the disease under the selective pressure of bortezomib- containing treatment regimens, representing branching evolution. It is to speculate whether previous investigations negating the existence of PSMB5 mutations in proteasome inhibitor treated patients might not have sequenced deep enough or did set up a sensitivity cut-off too rigid to detect such subclonal mutations. Our finding suggests that the mutations identified may contribute to the development of proteasome inhibitor resistance, emphasizing the need for more detailed genomic characterization of tumors, including minor subclonal mutation detection. Functional analysis of the mutations is ongoing and results will be presented at the meeting.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.