Abstract
Introduction: Multiple myeloma (MM) is characterized by the presence of monoclonal protein (M-protein) in serum and/or urine, clonal plasma cell accumulation in bone marrow, and related organ or tissue impairment. MM patients are monitored during and after therapy using immunoglobulin, M-protein and free light chain assays. Minimal residual disease (MRD) assessment of bone marrow samples from MM patients in complete remission (CR) using next-generation sequencing (NGS)-based technology has been shown to have prognostic value, and previous studies have demonstrated evidence of circulating myeloma cells in peripheral blood. In this prospective study, we compared the sensitivity of RNA- and DNA-based MRD assays in peripheral blood (PB) and bone marrow (BM) samples collected from MM patients.
Methods: Matched BM and PB samples were obtained from 50 MM patients in various stages of their disease. Diagnostic BM samples were used to identify clonal immunoglobulin sequences (i.e. clonotypes) unique to each MM patient. Matched BM and PB samples were then assessed for the presence of the MM clonotype. Using universal primer sets, we amplified immunoglobulin (IgH) variable (V), diversity (D), and joining (J) gene segments, the incomplete IgH-DJ rearrangement, and IgK receptors. Genomic DNA samples were assessed for all 3 receptors, while RNA samples were only assessed for the functional IgH-VDJ and IgK receptors. Amplified products were sequenced to obtain >1 million reads and analyzed using standardized algorithms for clonotype quantification. Myeloma-specific IgH, IgK, and IgH-DJ clonotypes were identified for each patient based on their high frequency at diagnosis. The presence of the myeloma clonotype was then assessed in BM cells (DNA), PB cells (DNA and RNA), and PB plasma (DNA) samples. Myeloma clonotype levels were calculated as previously described (Martinez-Lopez et al. 2014).
Results: High-frequency myeloma clonotypes were identified in the BM of all 50 patients. We performed a sequence-level assessment of each clonotype to determine whether it was also evaluable in RNA. Non-functional clonotypes (e.g. IgH-DJ and kappa deleting element clones, frameshift/nonsense mutations, nonfunctional V or J segments, loss of cysteine in CDR3) were excluded from the analysis. 37 evaluable RNA clonotypes were identified in 28 of the 50 patients (56%).
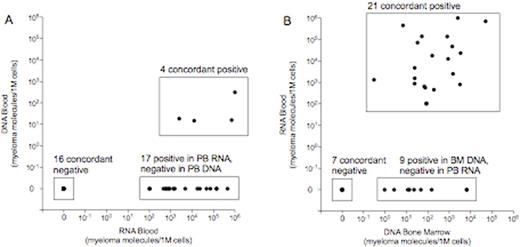
We compared the sensitivity of MRD assessment using DNA and RNA extracted from PB mononuclear cells. 20 clonotypes were qualitatively concordant in both DNA and RNA. 17 clonotypes were discordant, with all 17 clonotypes being detectable in RNA but not in DNA (Figure 1A). Only 4 of 21 (19%) clonotypes detectable in RNA were also detectable in DNA in the PB. Thus, RNA provides a clear sensitivity advantage over DNA analysis in PB cellular samples.
We then investigated whether the sensitivity advantage conferred by RNA in PB cells was equivalent or superior to the sensitivity of DNA analysis in BM cells. On a patient level, 23 patients were qualitatively concordant in the PB and BM assays. 5 patients were discordant, with all patients being positive in BM and negative in PB. 4 of the discordant patients were positive at low levels in the BM (1-10 MM clonotype molecules per 1 million cells). On a clonotype level, 28 clonotypes were qualitatively concordant in BM DNA and PB RNA. 9 clonotypes were discordant, with all 9 clonotypes being positive in BM DNA and negative in PB RNA (Figure 1B). Nevertheless, PB RNA detected MRD in 21 of 30 with measurable disease in the BM DNA (ie, 70% sensitivity). These results demonstrate that MRD assessment using PB RNA is more informative than PB DNA in MM; however, despite the sensitivity improvement provided by RNA, MRD assessment in the BM DNA remains the superior sample source for MRD assessment, particularly when achievement of MRD negativity is the goal of therapy.
Conclusions: NGS-based MRD assessment of BM in myeloma patients has been shown to have prognostic value. PB-based MRD assessment would improve clinical MRD assessment and monitoring paradigms. Our results demonstrate that RNA provides increased sensitivity for blood-based MRD assessment. Additional assay optimization to improve the fraction of clonotypes evaluable in RNA and to enhance sensitivity compared with BM is necessary before PB MRD monitoring in MM patients can be incorporated into routine clinical practice.
Wolf:Telomere Diagnostics: Consultancy; Pharmacyclics: Honoraria; Amgen: Honoraria; Takeda: Honoraria; Celgene: Honoraria. Kong:Adaptive Biotechnologies Corp: Employment, Equity Ownership. Bayes:Adaptive Biotechnologies Corp: Equity Ownership. Carlton:Adaptive Biotechnologies: Employment, Equity Ownership. Martin:Sanofi: Research Funding; Amgen: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.