Abstract
Introduction: The rapid technological advances in next generation sequencing (NGS) have allowed oncologists to sequence tumorexomesin a clinically meaningful period of time. NGS allows for identification of alterations that may be potentially targetable by already available Food and Drug Administration (FDA) approved drugs, providing a biologic rational for consideration of therapies and/or experimental treatments (clinical trials) that would not have otherwise been contemplated. Here we report our experience using NGS in a cohort of 60 patients with various lymphoid malignancies.
Methods: We retrospectively reviewed the medical charts of 60 patients with various lymphoid malignancies who had undergone NGS. Patients were seen at the UCSD Moores Cancer Center (La Jolla, CA) from October 2012 until March 2016. We collected tumor samples from tissue (Table 1) or peripheral blood from 60 patients that were submitted for testing to Foundation Medicine, a clinical laboratory improvement amendments (CLIA)-certified lab. Hybrid capture based NGS (405 gene panel) was performed (http://www.foundationone.com/). The methods used in this assay have been described in detail in a previous report (He, J. et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016. 127:3004-3014.).
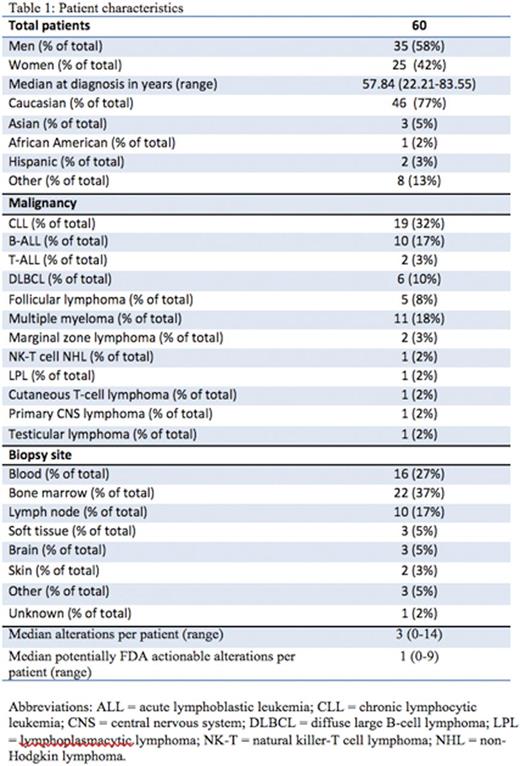
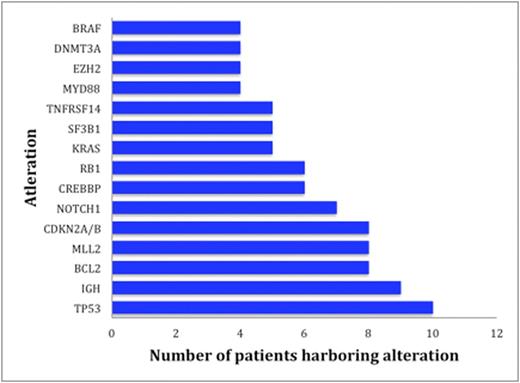
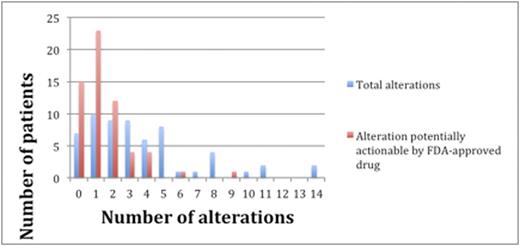
Results: Sixty patients including 35 men (58%) and 25 women (42%), were identified with lymphoid malignancies (Table 1). A total of 224 alterations were found by NGS in the entire cohort of 60 individuals. Types of alterations identified included substitutions, indels, copy number alterations (CNAs), and gene fusions. Figure 1 demonstrates the 15 most frequent alterations among the cohort: TP53 mutations (10 patients), IGH rearrangements (9 patients), loss of CDKN2A/B (8 patients), and BCL2 mutations (8 patients). The median number of alterations detected per patient was 3 (range, 0 to 14). Shown in Figure 2, 7 patients (12%) had no reportable alterations, 10 patients (17%) had 1 alteration, and 43 (71%) patients had 2 or more alterations. The maximum number of alterations identified was 14, which occurred in two patients (3%), one with chronic lymphocytic leukemia (CLL) and the other with diffuse large B-cell lymphoma (DLBCL). A total of 49 patients (82%) had potentially actionable alterations using FDA approved drugs and/or experimental therapies (clinical trials) while 11 patients (18%) had no theoretically actionable alterations. Only 3 patientshad an alteration for which an approved drug in the disease is available (on-label) while 45 patients (75%) had an alteration for which an approved drug is available in another disease (off-label). Twenty-three patients (38%), 12 patients (20%), and 10 patients (17%) had 1, 2 and³ 3 FDA targetable alterations, respectively. The median number of theoretical FDA actionable alterations was 1.
Conclusions:Most patients with lymphoid malignancies will have alterations that are potentially pharmacologically identified by NGS; however, only a minority of patients will have alterations for which an approved drug in the disease is available (on-label). Patients with lymphoid malignancies who have exhausted standard therapy or who are unable to tolerate chemotherapy may be excellent candidates for matched targeted therapies, ideally administered in the context of a clinical trial.
The blue bars represent the number of patients with the designated number of total alterations. The red bars represent the number of patients with the designated number of potentially actionable alterations by an FDA approved drug (on or off label).
The blue bars represent the number of patients with the designated number of total alterations. The red bars represent the number of patients with the designated number of potentially actionable alterations by an FDA approved drug (on or off label).
Goodman:Pfizer: Other: Fellowship funding. Costello:Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding. Kurzrock:Serono: Research Funding; Pfizer: Research Funding; Merck: Research Funding; Actuate Therapeutics: Research Funding; Sequenom: Research Funding; X Biotech: Research Funding; Genentech: Research Funding; Curematch: Equity Ownership; Novena: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.