Abstract
Introduction
We conducted a study to see the activity of hypomethylating agent (HMA) in relapsed refractory lymphoma. In parallel, we studied KIR expression and promoter methylation before and after treatment with HMA. Allele-specific stochastic KIR expression is maintained in mature natural killer (NK) cells by a combination of DNA methylation and histone modification, with DNA methylation being dominant. The aim of this part of the study is to understand how KIR gene expression changes in response to epigenetic therapy, so that we then can manipulate NK cells to optimize the effect of chemo-immunotherapy of neoplastic diseases.
Methods
Blood samples were collected from patients enrolled in an open-labeled phase I trial of the combination of azacitidine with cyclophosphamide, vincristine and rituximab in relapsed/refractory lymphoma.
Regimen: Azacitidine on days one through five (starting at 25mg/m2), followed by oral cyclophosphamide at 300 mg/m2 on days six through nine, vincristine 1.4 mg/m2 day eight, and rituximab 375 mg/m2 day eight. Each cycle was to be repeated every 21 days, up to eight cycles if participants continued to benefit from therapy.
Sampling: Between 10-20uL whole blood was collected from eligible patients before commencement of therapy and on Cycle 1 Day 5, Cycle 2 Day 1 and Cycle 3 Day 1. Peripheral blood mononuclear cells (PBMC) were isolated immediately ex vivo using Lymphocyte Separation Media (Lonza), and were divided into two aliquots: one for flowcytometry and the other for pyrosequencing (Epigendx, Hopkinton, MA).
Samples from each time point were stained with fluorescently-labelled antibodies to CD3, CD16, CD56, CD14, CD20 and four different KIRs. For calculating the number of KIRs per NK cell, we used a gating strategy in which CD56dim NK cells are identified, and then the number of KIR expression is counted on those by sequential gating. The percentages of monocytes and lymphocytes among the PBMC population were also determined using CD14 and CD20 antibodies. Monocytes were further characterized according to their CD16 expression into Classical (CD14highCD16negative), Intermediate (CD14highCD16low), and Non-classical (CD14lowCD16high).
Results
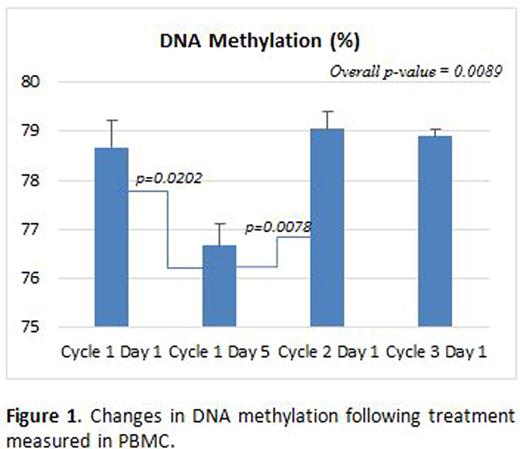
PBMC from 10 patients were available for flow cytometric analysis and pyrosequencing. There were statistically significant differences (p=0.0089) in percent of DNA methylation determined by PBMC pyrosequencing, with significant decrease from Cycle 1 Day 1 to Cycle 1 Day 5 and then an increase from Cycle 1 day 5 to Cycle 2 Day 1. The decline in the percent of DNA methylation in PBMC at day 5 of cycle 1 of the chemotherapy was also found to be statistically significant (Figure 1; p=0.0202).
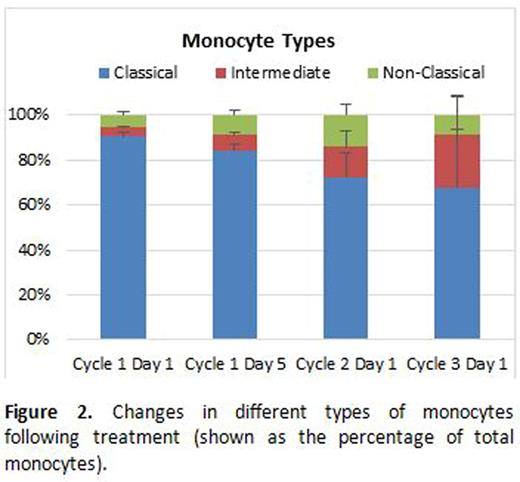
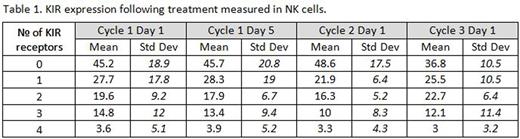
There was a large variation in the types and numbers of KIR molecules expressed at baseline (Table 1) on CD56dim NK cells among the patients in the cohort studied, with no significant differences between the various time points and response to treatment. Despite an apparent trend of increasing percentages of non-classical and intermediate monocytes (at the expense of classical monocytes), the data was not statistically significant (Figure 2; p=0.06). We also noticed a trend of decreasing percentage of monocytes and increasing percentage of lymphocytes in the PBMC population with increasing treatment cycles. However, this was not statistically significant either.
Conclusions
KIR expression may play role in HMA induced tumor cell killing. In our study, no significant changes were seen in the number of KIR receptors on NK cells post-treatment with hypomethylating agent despite observing a reduction in global DNA methylation in PBMC. This could be due to a number of reasons, including the small size of the study, the rapid turnover of NK cells, or that the reduced methylation was not sufficient to induce changes in KIR expression. The significance of the increase in the number of non-classical and intermediate monocytes following treatment with HMA is not well understood and needs further investigation.
(The study was supported by Celgene)
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.