Abstract
Biochemical aspects of cellular process are well characterized, but more recently, it has been shown that cells dynamically sense and respond to biophysical cues such as substrate stiffness and geometrical constraints; physical cues even direct cell differentiation and stem cell lineage (Discher et al, Science, 2005). In hematology, we know that platelets are shear activated and attenuate force based on substrate stiffness, and that endothelial cells align with flow and are activated by shear stress. Blood cells pass through, and interact with, biological matrices such as fibrin clots and the vascular wall, but the physical and biochemical aspects of these interactions are indistinguishable from one another in vivo. As such, there is a gap in knowledge as to how blood cells respond to matrices as they transit through them.
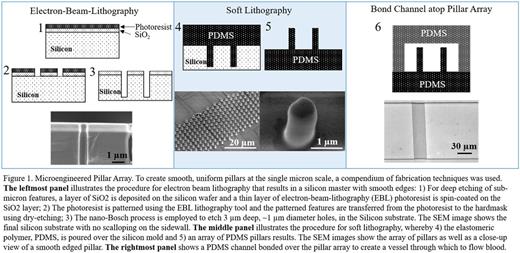
To decouple the physical and biochemical interactions of blood cells and biological matrices, we sought to recreate the physical geometry of a fibrin network in a controlled, non-biological, in vitro microfluidic system. To this end, we designed a two-part microfluidic device comprised of an array of micron sized pillars (~1 µm diameter, 3 µm height, and 2 µm gap between pillars) overlaid with a microfluidic channel (Fig 1). The dimensions of the pillars are on the order of the diameter of fibrin fibers and the mesh size of a fibrin gel (Okada et al, J. Biol. Chem, 1985), while channels of various dimensions can be bonded over the pillar array to represent various biological scenarios. Standard microfluidic processes cannot produce pillars with the feature sizes reported herein, so electron beam lithography was used to create the mold from which the elastomeric pillars are made.
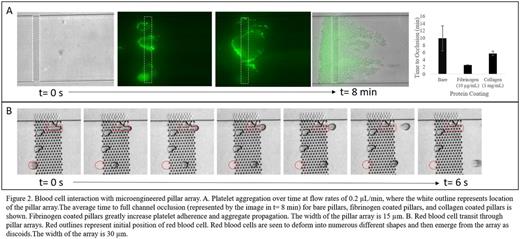
The biophysical interaction of platelets flowing through fibrin mesh (absent biological factors) was recreated by a pillar array oriented perpendicular to the direction of flow in a 6 µm tall channel. When washed platelets are perfused through the system, they adhere to the pillars, aggregate, and form an occlusive mass that extends to the edges of the array (Fig 2A). Platelet adhesion initiates exclusively at the pillars and aggregation propagates to the extents of the channel area perpendicular to flow, resulting in channel occlusion and flow cessation. These findings show that in the absence of platelet agonists and biological ligands, platelets are activated by the shear environment afforded by the presence of fibrin fibers. Thus, in addition to the biochemical players in clot formation, the geometry of the fibrin mesh plays a role in platelet adhesion, and clot propagation. As expected, passive adsorption of fibrinogen and collagen to the pillar surfaces enhances platelet aggregation, as evidenced by a decrease in time to channel occlusion from 10 min to 2 min and 6 min, respectively. With thus we see the synergistic effect of biophysical and biochemical factors in clot propagation. This novel microfluidic system both separates biophysical and biochemical aspects of clot formation and allows researchers to specify the precise location and extent of clot formation in vitro.
Platelets are not the only blood cells to interact with and react to physical barriers. Red blood cell (RBC) deformation has been historically studied in single cell assays, SEM studies of fixed clots, and more recently after RBCs have passed through a filtration system comprised of either beads or long slits (Deplaine et al, Blood, 2010); however, real time visualization of RBC deformation in geometries representative of biological matrices has remained elusive. The deformation (and possible fragmentation) that RBCs undergo when passing through the physical challenges of a fibrin matrix or the interendothelial slits of the spleen can be visualized in our system: an array of pillars overlaid by a 3 µm channel. Our findings visually suggest that red blood cells are able to deform through the matrix with little effect on their membranes, and that exposure to high shear gradients alone does not cause cell fragmentation (Figure 2B). The ready deformation and transit of healthy RBCs in our system confirms recent computational studies of RBC filtration by the spleen (Pivkin et al, PNAS, 2016). Further studies will give insight into the deformation and transit of sickled cells and malaria infected RBCs in physical matrices. Overall, our microfluidic studies give novel insight into the biophysical aspect of blood cell interactions with biological matrices.
Lam:Sanguina, LLC: Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.