Key Points
A total of 4516 lncRNAs were identified across multiple stages of B-cell development and activation.
Abstract
Long noncoding RNAs (lncRNAs) are potentially important regulators of cell differentiation and development, but little is known about their roles in B lymphocytes. Using RNA-seq and de novo transcript assembly, we identified 4516 lncRNAs expressed in 11 stages of B-cell development and activation. Most of these lncRNAs have not been previously detected, even in the closely related T-cell lineage. Comparison with lncRNAs previously described in human B cells identified 185 mouse lncRNAs that have human orthologs. Using chromatin immunoprecipitation-seq, we classified 20% of the lncRNAs as either enhancer-associated (eRNA) or promoter-associated RNAs. We identified 126 eRNAs whose expression closely correlated with the nearest coding gene, thereby indicating the likely location of numerous enhancers active in the B-cell lineage. Furthermore, using this catalog of newly discovered lncRNAs, we show that PAX5, a transcription factor required to specify the B-cell lineage, bound to and regulated the expression of 109 lncRNAs in pro-B and mature B cells and 184 lncRNAs in acute lymphoblastic leukemia.
Introduction
Long noncoding RNAs (lncRNAs) have emerging roles in innate and adaptive immunity. For example, lnc-DC is required for normal dendritic cell differentiation and function,1 IL1β-eRNA and IL1β-RBT46 are required for lipopolysaccharide-induced pro-inflammatory responses in monocytes,2 and NRAV modulates cellular responses to viral infections.3 In T cells, an intronic lncRNA NRON abrogates the nuclear transport of nuclear factor of activated T cells, and hence modulates expression of interleukin-2.4 In B-cell lymphomas, the lncRNA Fas-AS1 modulates expression of soluble Fas receptor messenger RNA, an important regulator of apoptosis.5 Thus, lncRNAs have the potential to influence both normal and pathological immune cell development and function.
LncRNAs may operate via a variety of molecular mechanisms.6 For example, enhancer-associated lncRNAs (eRNAs) act in cis and originate from transcribed extragenic or intragenic enhancer regions, whereas promoter-associated lncRNAs (pRNAs) can act in trans and originate from canonical promoter-derived transcriptional activity.7,8 These 2 broad lncRNA categories are distinguished by the ratio of mono- vs tri-methylation of histone 3 lysine 4 (H3K4me1/H3K4me3).8 Compared with pRNAs, eRNAs tend to exhibit more restricted expression and their RNA sequences show less constraint.8
Advances in sequencing technology have enabled the identification of large numbers of putative lncRNA loci.9,10 However, the proportion of lncRNAs with clearly defined function is small,11,12 caused in part by poor annotation of lncRNAs expressed in a tissue of interest, making it difficult to select candidate lncRNAs for targeted studies. This is a consequence of the expression patterns of lncRNAs, which are often restricted to 1 or very few tissues or cell types.9 Recent studies have addressed this limitation by surveying lncRNA expression in several organisms and tissues,13-17 including murine T cells18 ; however, there have been no comparable attempts to use sequencing technologies to describe the murine B-cell lncRNA repertoire.
To facilitate the study of lncRNA biology in B cells, we describe a catalog of 4516 de novo assembled high-confidence lncRNAs expressed in 11 mouse B-cell populations. We identify human lncRNAs that may be orthologs of the mouse genes. Furthermore, we classify subsets of eRNAs and pRNAs and perform an unsupervised clustering analysis to associate lncRNAs with messenger RNAs at key stages of B-cell development. Finally, we use the lncRNA catalog to show that PAX5, a transcription factor required to specify the B-cell lineage,19 binds to and regulates expression of lncRNA loci in both pro-B and mature B cells as well as in acute lymphoblastic leukemia.
Materials and methods
Mice
All RNA-seq and chromatin immunoprecipitation (ChIP)-seq experiments were performed with female C57BL/6JNimr mice aged 7-9 weeks, except for RNA-seq of plasmablasts and plasma cells, which were obtained from 12- to 14-week-old Blimp1-GFP mice.20
Cell sorting
Gating strategies for cell sorting are shown in supplemental Figure 1, available on the Blood Web site.
RNA-seq
Sorted populations of cells were resuspended in Trizol (Life Technologies), and RNA was purified using the RNeasy Mini Kit (Qiagen). RNA quality was assessed using the 2100 Expert Agilent Bioanalyzer. For all samples except plasmablasts and plasma cells, stranded polyA-enriched libraries were made using the Stranded TruSeq RNA Sample Preparation Kit (Illumina) and sequenced on the HiSeq 2500 (Illumina), collecting 100 base paired-end reads. For plasma cells and plasmablasts, unstranded non-ribosomal RNA–enriched libraries were made using the SMARTer Ultra Low Input RNA Kit for Sequencing v3 (Clontech) and sequenced collecting 50 base paired-end reads.
ChIP-seq
ChIP immunoprecipitation-sequencing was performed in triplicate for all stages of B-cell development, except plasmablasts and plasma cells, as described previously.21 For details, see the supplemental Methods.
RNA-seq read alignment and transcript assembly
RNA-seq reads were aligned to the C57BL/6J mouse reference genome (mm10, GRCm38) using STAR22 v2.3.0e. Transcript assembly was performed separately for all samples except plasmablasts and plasma cells using Cufflinks23 v2.2.0. Nonuniquely mapping reads were retained during assembly and potential transcripts (transfrags) were discarded when they contained fewer than 5 successfully mapped reads. Individual assemblies were subsequently compared and transfrags were discarded if they were not assembled in at least 2 samples.
Identification of lncRNAs
The lncRNA discovery pipeline is depicted in supplemental Figure 2A. Following assembly, transcripts <200 bp in length were discarded. Remaining transcripts were filtered against existing databases (Ensembl v72 and National Center for Biotechnology Information RefSeq) and discarded if they intersected (≥1 bp, on the same strand) intervals annotated as anything other than noncoding RNA. Transcripts were also discarded if they intersected nuclear mitochondrial DNA or pseudogenes predicted using Exonerate,24 if they were in close proximity downstream of a protein-coding gene, or if they were classified as coding by both Coding Potential Calculator25 and PhyloCSF.26 Further details, including a description of lncRNA nomenclature, are provided in supplemental Methods. LncRNAs classed as intergenic (>5 kb from a protein-coding gene) were identified as eRNAs or pRNAs using H3K4me1 and H3K4me3 ChIP-seq data in conjunction with a stringent classification pipeline (supplemental Figure 2B), in which successfully classified loci were required to intersect a called chromatin peak and to show 4-fold greater coverage of reads arising from their characteristic chromatin mark.
UCSC public hub
Genomic data has been visualized as a UCSC public hub, accessible at https://www.cgat.org/downloads/public/projects/proj010/UCSC_track_hub/hub.txt.
Results
A total of 4516 lncRNA loci expressed during B-cell development
To identify and characterize lncRNAs expressed during B-cell development and activation, we used fluorescence-activated cell sorting to obtain 5 replicate cell samples for each of 8 purified B-cell populations derived from the bone marrow, spleen, and peritoneal cavity of adult female C57BL/6JNimr mice (Figure 1A; supplemental Figure 1A-D). These populations represent the major subsets of developing B cells in the bone marrow (pro-B cells, pre-B cells, and immature B cells) and subsets of mature B cells in the bone marrow, spleen (follicular and marginal zone B cells), and peritoneal cavity (B1a cells). In addition, we also sorted germinal center (GC) B cells, an activated B-cell subset. From each of these 40 samples, we extracted and sequenced the polyA+ RNA fraction in a strand-specific manner to an average depth of 42 million uniquely mapped reads, using a 2 × 100 base paired-end sequencing protocol.
Identification of lncRNAs expressed in B cells. (A) Schematic representation of the ontogenetic relationships between B-cell populations used to generate the lncRNA catalog. Solid arrows indicate developmental progression through B-cell stages or activation (to GC B cells). Dashed line indicates recirculation of follicular B cells back to the bone marrow. B1A, B1a B cells; FO, follicular B cells; GC, germinal center B cells; IMM, immature B cells; MAT, mature B cells; MZ, marginal zone B cells; PRE, pre-B cells ; PRO, pro-B cells. (B) Genomic distribution of the 1491 multiexon and 3025 single-exon lncRNAs identified by this study. Positions are described relative to Ensembl v72 protein-coding gene annotations as antisense (overlapping a coding gene on antisense strand), flanking (<5 kb from coding gene), and intergenic (>5 kb from coding gene). (C) Overlap between the 2349 intergenic lncRNAs identified by this study (B cell), with those identified in T lineage cells18 (T cell), and those annotated in Ensembl v78. Kernel density plots representing the distribution of distance between each multiexon (D) and single-exon (E) intergenic lncRNA TSS and the nearest annotated TSS on the same strand that appeared in 2 or more of the 128 mouse cell lines considered by the FANTOM5 consortium. CAGE, cap analysis of gene expression. Shaded regions indicate a null distribution as measured by distance to the nearest FANTOM5 annotated TSS on the opposing strand. Vertical gray dashed line indicates a distance of 500 bp. (F) Coverage of the genome and of lncRNA exons by the indicated transposon elements.
Identification of lncRNAs expressed in B cells. (A) Schematic representation of the ontogenetic relationships between B-cell populations used to generate the lncRNA catalog. Solid arrows indicate developmental progression through B-cell stages or activation (to GC B cells). Dashed line indicates recirculation of follicular B cells back to the bone marrow. B1A, B1a B cells; FO, follicular B cells; GC, germinal center B cells; IMM, immature B cells; MAT, mature B cells; MZ, marginal zone B cells; PRE, pre-B cells ; PRO, pro-B cells. (B) Genomic distribution of the 1491 multiexon and 3025 single-exon lncRNAs identified by this study. Positions are described relative to Ensembl v72 protein-coding gene annotations as antisense (overlapping a coding gene on antisense strand), flanking (<5 kb from coding gene), and intergenic (>5 kb from coding gene). (C) Overlap between the 2349 intergenic lncRNAs identified by this study (B cell), with those identified in T lineage cells18 (T cell), and those annotated in Ensembl v78. Kernel density plots representing the distribution of distance between each multiexon (D) and single-exon (E) intergenic lncRNA TSS and the nearest annotated TSS on the same strand that appeared in 2 or more of the 128 mouse cell lines considered by the FANTOM5 consortium. CAGE, cap analysis of gene expression. Shaded regions indicate a null distribution as measured by distance to the nearest FANTOM5 annotated TSS on the opposing strand. Vertical gray dashed line indicates a distance of 500 bp. (F) Coverage of the genome and of lncRNA exons by the indicated transposon elements.
To identify lncRNAs, we used a de novo transcript assembly pipeline (supplemental Figure 2A). Stringency of selection of potential lncRNA loci was assured by discarding transcripts whose assembly was not replicated in at least 2 of the 40 sample libraries, by discarding loci represented in public databases of protein-coding annotations (Ensembl and RefSeq), and by discarding loci containing transcripts with predicted coding potential (supplemental Figure 3A-C). Using this process, we identified 4516 lncRNA loci that were expressed in at least 1 of the 8 B-cell populations, and comprised, on average, 1.8 transcripts per locus (supplemental Table 1). To validate the assembly process, we used polymerase chain reaction to check for expression of predicted lncRNAs. Of 53 lncRNAs with expression values ranging between 0.9 and 667 fragments per kilobase per million (FPKM), we were able to successfully validate 47 (89%), demonstrating that the assembly pipeline had generated reliable transcripts (supplemental Figure 4).
Of the predicted lncRNA loci, 3025 (67%) showed no evidence of splicing (single-exon loci), whereas the remaining 1491 showed evidence of at least 1 spliced transcript (multiexon loci). Approximately half of all single and multiexon loci were intergenic, with the remainder being either flanking (<5 kb from a protein-coding gene) or overlapping a protein-coding gene on the antisense strand (Figure 1B; supplemental Table 2). The single-exon and multiexon lncRNAs showed similar coding potential, expression levels and mean exon sizes, supporting the view that they belong to the same gene class (supplemental Figure 5A-D).
Intergenic B-cell lncRNAs show little overlap with existing lncRNA catalogs
We compared the 2349 intergenic lncRNA loci identified in this study with intergenic lncRNAs reported in mouse T-cell subsets,18 and with the latest Ensembl (v74) long intergenic noncoding RNA annotations. A total of 1829 (78%) of the intergenic lncRNAs in our B-cell catalog did not overlap on the relevant strand with either the T cell or Ensembl long intergenic noncoding RNAs (Figure 1C), reinforcing the notion that many lncRNAs are tissue- or cell type–specific in their expression.9,12,18
The highly tissue-specific expression of B-cell lncRNAs was further confirmed by comparison of our catalog with the Functional Annotation of the Mammalian Gene 5 (FANTOM5) study of transcriptional start sites (TSS) in 128 mouse primary cell types,27 which included a single B-cell sample. Because FANTOM annotation required TSS to be detected in 2 or more samples, we did not expect this dataset to include TSS for lncRNAs expressed only in B cells. Accordingly, only 299 (13%) of our intergenic lncRNAs were within 500 bp of a FANTOM-annotated TSS (Figure 1D-E). Notably, a greater overlap with FANTOM-annotated TSS was observed for multiexon transcripts than for single-exon transcripts. The same trend was observed in comparisons with previous lncRNA annotations (supplemental Figure 5E-F). We therefore conclude that expression at an lncRNA locus is more likely to be recapitulated in different cell types if the locus contains evidence of splicing.
A previous study had shown that many lncRNAs overlap with transposable elements (TEs).28 In agreement with this, we find that approximately 40% of the lncRNAs identified here overlap with TEs, and a comparison of these with the genomic content of TEs shows an enrichment of endogenous retroviruses and a relative depletion of LINE elements, similar to the previous report (Figure 1F; supplemental Table 3).
In summary, we have generated a novel, high-confidence catalog of lncRNAs expressed across key stages of B-cell development. To facilitate the use of this catalog, we have visualized our genomic data as a UCSC public hub.
Chromatin signatures identify intergenic lncRNAs with enhancer-associated and promoter-associated expression
To identify intergenic lncRNAs within our catalog that may be either eRNAs or pRNAs, we determined the genome-wide abundance of H3K4me1 and H3K4me3 chromatin marks known to distinguish these 2 lncRNA subtypes8 (Figure 2A-B). We quantified the relative abundance of these marks at the proposed TSS of 2349 intergenic lncRNAs across all 8 B-cell populations, and identified eRNAs and pRNAs using stringent selection criteria (supplemental Figure 2B).
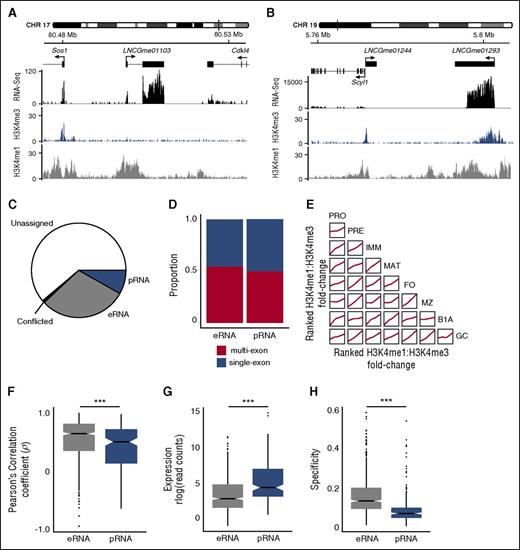
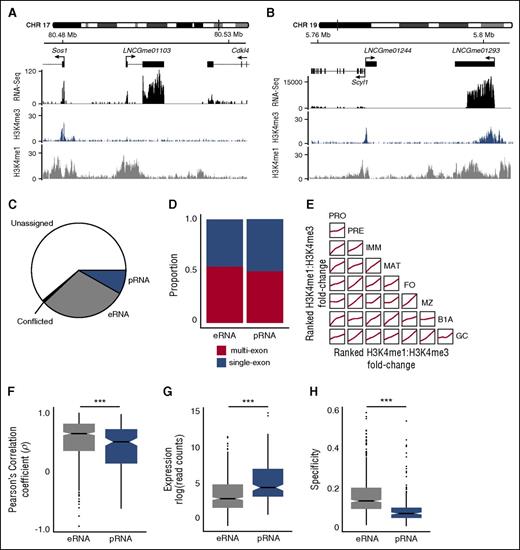
Identification of intergenic lncRNAs with enhancerlike and promoterlike characteristics. Examples of intergenic lncRNA loci with chromatin signatures in marginal zone B cells that are characteristic of (A) eRNA loci (LNCGme01103) and (B) pRNA loci (LNCGme01293). The former are distinguished by high H3K4me1 read coverage across the TSS and the absence of a corresponding H3K4me3 peak. The latter are distinguished by high H3K4me3 coverage, the presence of which excludes H3K4me1 from the TSS. (B) Also shown is a second lncRNA (LNCGme01244) that arises as a result of bidirectional transcription from the Scyl1 promoter, but this is not classified as an eRNA or pRNA because its proximity to a coding gene. CHR, chromosome. (C) The proportion of the 2349 intergenic lncRNAs identified in this study that could be classified as either eRNAs or pRNAs on the basis of their chromatin state. Remaining lncRNAs are either classified as unassigned (insufficient read coverage/fold-change to determine chromatin state) or conflicted (classified as eRNA in 1 B-cell stage and pRNA in another). (D) The proportion of eRNA and pRNA loci that are classified as multiexon or single exon. (E) Pairwise comparisons showing the consistency of chromatin signatures across B-cell populations. Within each B-cell population, intergenic lncRNAs are ranked on the ratio of H3K4me1:H3K4me3 coverage across their TSS. Individual plots show the local regression (loess) of rank order between 2 B-cell populations. (F) Distribution of the Pearson’s correlation coefficient between the expression of an eRNA or pRNA and expression of the more highly correlated of either its nearest upstream or downstream protein-coding gene. (G) Distribution of median expression values (rlog-transformed read counts) calculated across all cell stages. (H) Distribution of cell stage specificity of expression of eRNAs and pRNAs. ***Mann-Whitney U test: p < .0001.
Identification of intergenic lncRNAs with enhancerlike and promoterlike characteristics. Examples of intergenic lncRNA loci with chromatin signatures in marginal zone B cells that are characteristic of (A) eRNA loci (LNCGme01103) and (B) pRNA loci (LNCGme01293). The former are distinguished by high H3K4me1 read coverage across the TSS and the absence of a corresponding H3K4me3 peak. The latter are distinguished by high H3K4me3 coverage, the presence of which excludes H3K4me1 from the TSS. (B) Also shown is a second lncRNA (LNCGme01244) that arises as a result of bidirectional transcription from the Scyl1 promoter, but this is not classified as an eRNA or pRNA because its proximity to a coding gene. CHR, chromosome. (C) The proportion of the 2349 intergenic lncRNAs identified in this study that could be classified as either eRNAs or pRNAs on the basis of their chromatin state. Remaining lncRNAs are either classified as unassigned (insufficient read coverage/fold-change to determine chromatin state) or conflicted (classified as eRNA in 1 B-cell stage and pRNA in another). (D) The proportion of eRNA and pRNA loci that are classified as multiexon or single exon. (E) Pairwise comparisons showing the consistency of chromatin signatures across B-cell populations. Within each B-cell population, intergenic lncRNAs are ranked on the ratio of H3K4me1:H3K4me3 coverage across their TSS. Individual plots show the local regression (loess) of rank order between 2 B-cell populations. (F) Distribution of the Pearson’s correlation coefficient between the expression of an eRNA or pRNA and expression of the more highly correlated of either its nearest upstream or downstream protein-coding gene. (G) Distribution of median expression values (rlog-transformed read counts) calculated across all cell stages. (H) Distribution of cell stage specificity of expression of eRNAs and pRNAs. ***Mann-Whitney U test: p < .0001.
A total of 702 eRNAs and 192 pRNAs were predicted in 1 or more B-cell populations (Figure 2C; supplemental Figure 6A-C; supplemental Table 2) and were evenly distributed between single-exon (320 eRNAs, 96 pRNAs) and multiexon loci (382 eRNAs, 96 pRNAs, Figure 2D). H3K4me1 and H3K4me3 marks at all lncRNA loci remained broadly consistent across B-cell populations (Figure 2E). Furthermore, lncRNA classified as eRNAs or pRNAs in 1 B-cell population displayed a comparable chromatin signature in other cell populations (supplemental Figure 6D-E), suggesting that these classifications represent 2 largely mutually exclusive lncRNA classes.
In agreement with earlier studies,8 B-cell eRNAs were distinguishable from pRNAs by a significantly higher correlation of expression with their proximal protein-coding gene (Figure 2F), by lower expression (Figure 2G), and by expression in fewer cell subsets (Figure 2H). Thus, lncRNAs with a high H3K4me1:H3K4me3 ratio at their TSS show characteristics typical of eRNAs and their expression tends to be associated with the expression of adjacent protein-coding genes.
LncRNAs are expressed at discrete stages during B-cell development
Previous studies in the T-cell lineage had shown that intergenic lncRNA expression is highly restricted even between closely related T-cell subsets.17,18 If this were also the case for B cells, then B-cell subsets should be distinguishable on the basis of lncRNA expression. Indeed, using both hierarchical clustering and principal component analysis on either coding gene or lncRNA expression, all 8 B-cell populations were distinguishable and replicate samples clustered closely (Figure 3A-B; supplemental Figure 7A-D). For both types of genes, follicular and marginal zone B cells were the most closely related, whereas B1a cells and GC B cells showed the greatest distinction from other cell types.
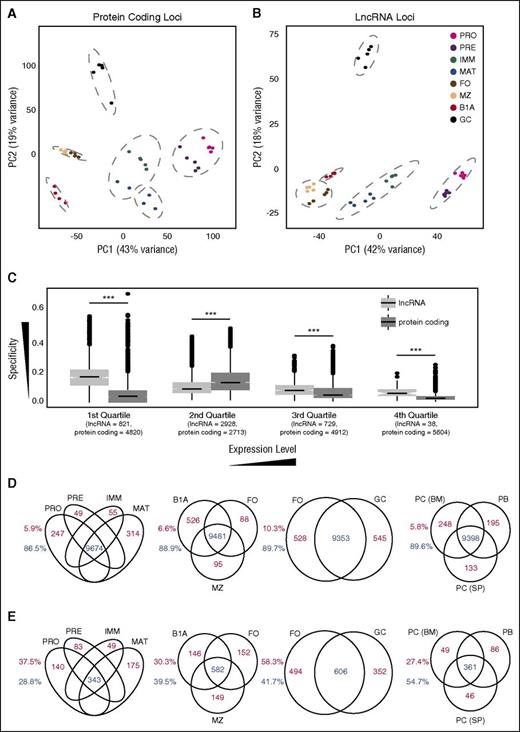
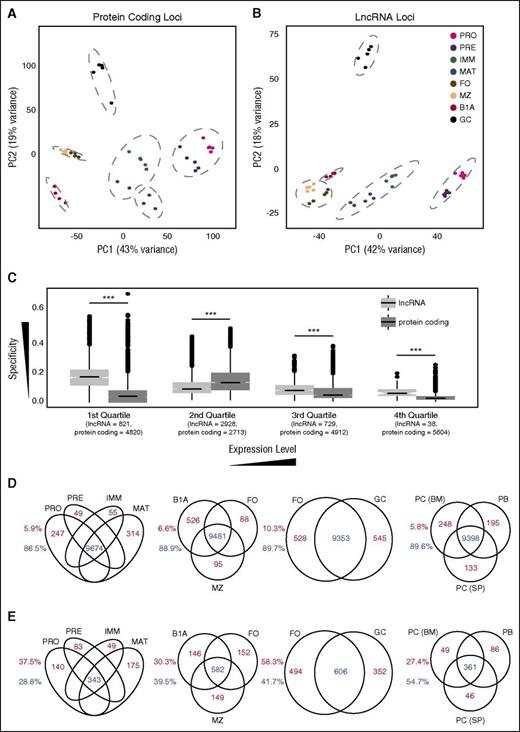
Cell stage–specific expression at lncRNA and protein-coding loci. Principal component analysis of regularized log-transformed expression patterns at (A) protein-coding loci and (B) the 4516 lncRNA loci identified in this study. Dashed gray lines indicate groups identified by unsupervised hierarchical clustering (Supplemental Figure 7A-B). (C) Box plots of specificity of expression of all coding and lncRNA loci separated into quartile bins on the basis of their median expression across all 8 B-cell stages. Numbers below each quartile indicate the number of lncRNA and protein-coding loci that fall into each category. Venn diagrams showing the number of protein-coding (D) and lncRNA (E) loci that are either expressed in multiple cell populations (blue) or expressed in a single cell population (red) at an FPKM threshold of 1.0. Splenic plasmablasts (PB) and plasma cells (PC (SP)), and bone marrow plasma cells (PC (BM)). Numbers adjacent to each plot depict the proportion of loci falling into each category.
Cell stage–specific expression at lncRNA and protein-coding loci. Principal component analysis of regularized log-transformed expression patterns at (A) protein-coding loci and (B) the 4516 lncRNA loci identified in this study. Dashed gray lines indicate groups identified by unsupervised hierarchical clustering (Supplemental Figure 7A-B). (C) Box plots of specificity of expression of all coding and lncRNA loci separated into quartile bins on the basis of their median expression across all 8 B-cell stages. Numbers below each quartile indicate the number of lncRNA and protein-coding loci that fall into each category. Venn diagrams showing the number of protein-coding (D) and lncRNA (E) loci that are either expressed in multiple cell populations (blue) or expressed in a single cell population (red) at an FPKM threshold of 1.0. Splenic plasmablasts (PB) and plasma cells (PC (SP)), and bone marrow plasma cells (PC (BM)). Numbers adjacent to each plot depict the proportion of loci falling into each category.
A greater proportion of B-cell lncRNAs was differentially expressed between cell populations than protein-coding genes (supplemental Figure 8A). Although cell type specificity was, as expected, dependent on expression level, even at equivalent expression levels, lncRNAs displayed greater cell-type restriction than coding genes (Figure 3C; supplemental Figure 8B-C), suggesting that lncRNAs have very restricted spatiotemporal roles during B-cell development.
The trend for lncRNAs to be more restricted in expression than protein-coding genes was observed across all stages of B-cell development. At a defined expression threshold (FPKM >1), a greater proportion of protein-coding genes than lncRNAs were expressed in all bone marrow–derived B-cell populations, in all splenic and peritoneal cavity mature B-cell populations, and in both naïve and activated B-cell populations (Figure 3D-E).
To extend this analysis further, we examined expression of the lncRNAs in antibody-secreting cells. We performed RNA-seq on splenic plasmablasts and plasma cells and on bone marrow plasma cells (supplemental Figure 1E-F; supplemental Figure 7E) and determined expression of lncRNAs and coding genes in these 3 additional subsets (supplemental Table 2). Principal component analysis showed that the 3 subsets clustered closely together (supplemental Figure 7F-G) and lncRNAs were again more cell type–specific (Figure 3D-E).
Generating hypotheses for lncRNA functions through correlation with protein-coding gene expression
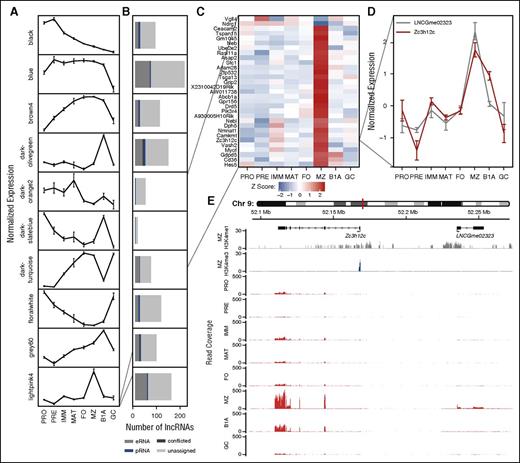
One genome-scale approach to generate hypotheses regarding lncRNA function is to exploit a “guilt by association” approach by correlating expression of lncRNAs with coding genes of known biological function.9,29 We predicted the biological processes to which lncRNAs might contribute by clustering protein-coding loci expressed in at least 1 B-cell type (median FPKM >1) into 1 of 17 clusters, based on correlation of expression across B-cell development30 (supplemental Figure 9A-B; supplemental Table 2). We subsequently correlated individual lncRNAs with the representative gene expression profile (eigengene) for each protein-coding cluster (Figure 4A), and defined an association between an lncRNA and a cluster of protein-coding genes at a Pearson correlation coefficient of |ρ| > 0.8 (Figure 4B). At this threshold, 987 (22%) lncRNAs associated with protein-coding gene clusters, suggesting that these lncRNAs contribute to the same biological processes as the protein-coding genes within the respective cluster. Although some clusters were characteristic of individual cell stages (Figure 4A), Gene Ontology enrichment analysis31 identified others that were associated with cellular processes (supplemental Table 4). For example, the lightpink4 cluster contains genes up- or downregulated in marginal zone B cells, whereas the floralwhite cluster is enriched for genes associated with the cell cycle, and its eigengene is highest in pro-B cells, pre-B cells, and GC B cells, the 3 subsets with the highest rates of cell division.
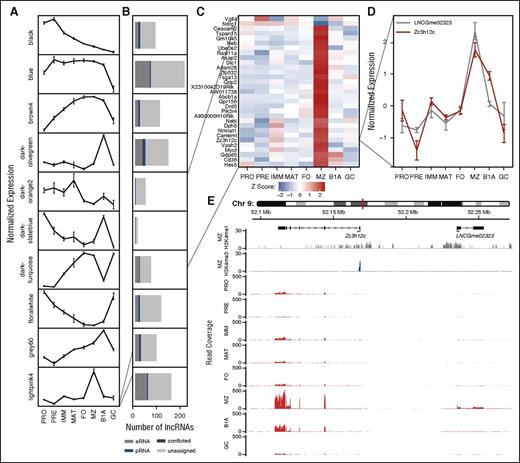
Association between lncRNAs and protein-coding genes based on correlation of expression across B-cell development. (A) WGCNA identifies clusters of protein-coding genes with comparable expression profiles across B-cell development: plots show a representative expression profile (eigengene) for each of 10 protein-coding gene clusters. (B) Stacked bar charts showing the number of lncRNAs whose expression is strongly correlated (|ρ| > 0.8) with the adjacent eigengene. Colors depict lncRNA classification on the basis of chromatin state. (Results are shown for WGCNA clusters with >10 associated lncRNAs.) (C) Heat map showing the normalized expression of cluster lightpink4 containing 31 protein-coding genes identified as upregulated or downregulated in marginal zone B cells. (D) The normalized expression profile of a single gene (Zc3h12c) from this cluster identified as upregulated in marginal zone B cells and a single eRNA (LNCGme02323) identified as strongly correlated with the respective WGCNA cluster. (E) Genome plots showing the location of Zc3h12c and LNCGme02323 as well as H3K4me1 and H3K4me3 chromatin signatures in marginal zone B cells and RNA-seq read coverage in all 8 B-cell populations considered in this study.
Association between lncRNAs and protein-coding genes based on correlation of expression across B-cell development. (A) WGCNA identifies clusters of protein-coding genes with comparable expression profiles across B-cell development: plots show a representative expression profile (eigengene) for each of 10 protein-coding gene clusters. (B) Stacked bar charts showing the number of lncRNAs whose expression is strongly correlated (|ρ| > 0.8) with the adjacent eigengene. Colors depict lncRNA classification on the basis of chromatin state. (Results are shown for WGCNA clusters with >10 associated lncRNAs.) (C) Heat map showing the normalized expression of cluster lightpink4 containing 31 protein-coding genes identified as upregulated or downregulated in marginal zone B cells. (D) The normalized expression profile of a single gene (Zc3h12c) from this cluster identified as upregulated in marginal zone B cells and a single eRNA (LNCGme02323) identified as strongly correlated with the respective WGCNA cluster. (E) Genome plots showing the location of Zc3h12c and LNCGme02323 as well as H3K4me1 and H3K4me3 chromatin signatures in marginal zone B cells and RNA-seq read coverage in all 8 B-cell populations considered in this study.
We previously showed an increased correlation of expression between eRNAs and their proximal protein-coding gene compared with pRNAs (Figure 2F); the association of lncRNAs with clusters of protein-coding genes allowed us to test this hypothesis further. Permutation tests demonstrated that eRNAs showed strongest correlation with the cluster containing their proximal protein-coding gene (P = .04), whereas no such trend was observed for pRNAs (P = .51).
Combining information about genomic location, chromatin state, and expression allowed us to identify eRNAs that may function in cis with genes of distinctive biological function. For example, Figure 4C shows a single cluster of protein-coding genes that exhibit large changes of expression within marginal zone B cells. Of the lncRNAs correlated with this cluster, we show 1 (LNCGme02323) that is proximal to cluster member Zc3h12c, encoding a zinc finger protein implicated in the regulation of pro-inflammatory activation in macrophages.32 Using our catalog, LNCGme02323 is identifiable as an eRNA that is coexpressed with Zc3h12c in marginal zone B cells and that lies ∼67 kb upstream of the coding gene TSS (Figure 4D-E), suggesting that this lncRNA may be transcribed from an enhancer controlling the expression of Zc3h12c. In total, we were able to identify 126 eRNA loci whose expression was most strongly correlated with that of a weighted gene coexpression network (WGCNA) module containing an adjacent protein-coding gene, including 48 with a correlation of |ρ| > 0.8 (supplemental Table 5).
Human lncRNA orthologs
To extend the utility of this catalog of mouse lncRNAs, we sought to identify potential orthologous human lncRNAs by comparing the mouse genes with lncRNAs previously identified in human B cells.17,33-35 Based on pairwise genomic alignments, we identified 185 mouse lncRNAs that have a human syntenic ortholog (supplemental Table 6). Taking advantage of the fact that eRNAs are a distinct class of lncRNA throughout B-cell development (supplemental Figure 6D), we compared mouse eRNAs with human eRNAs reported in CD19+ B cells.34 Pairing eRNAs adjacent to orthologous human and mouse coding genes identified a further 228 mouse eRNAs that have 1 or more human eRNA counterparts (supplemental Table 6). Such conserved location indicates that the human/mouse pairs may be marking enhancers of orthologous coding genes.
PAX5 regulates lncRNA expression during normal and malignant B-cell development
The transcription factor PAX5 is expressed throughout B-cell development and is crucial for commitment to the B-cell lineage as well as for maintaining B-cell homeostasis.19,36 PAX5 mediates these functions through the regulation of protein-coding gene expression, but it may also regulate lncRNA expression. Using previously published PAX5 binding sites,36 we identified 784 and 717 lncRNAs in pro-B cells and mature B cells, respectively, whose loci either overlapped or were <1 kb downstream of a PAX5 binding site that had no comparable association with a protein-coding gene. This represented a statistically significant enrichment compared with a null model of PAX5 binding (pro-B cells: 6.1-fold, P < 10−4; mature B cells: 10.5-fold, P < 10−4). Using published RNA-seq data for wild-type or Pax5-deficient pro-B cells or mature B cells,36 we further identified 28 and 87 of these lncRNA loci, respectively, for which there is evidence that PAX5 both binds and regulates their expression (supplemental Figure 10A-D; supplemental Table 7).
Disruption of PAX5 binding during early B-cell development is associated with development of B-progenitor acute lymphoblastic leukemia (B-ALL).37 For this reason, we extended our analysis of PAX5 binding and lncRNA expression using RNA-seq data from a mouse model of B-ALL in which Pax5 expression can be regulated by doxycycline.38 Induction of Pax5 expression within leukemic cells causes the tumor cells to differentiate and the leukemia to regress. Comparison of B-ALL cells with and without doxycycline-induced Pax5 expression revealed 331 differentially expressed lncRNAs (q < 0.05), of which a large majority (91%) were upregulated as a result of Pax5 expression (supplemental Table 7). Furthermore, most (184) of these lncRNAs originated from loci bound by PAX5 in pro-B cells or mature B cells (Figure 5A-B). As an example, we show 2 previously annotated lncRNA loci (LNCGme00432 and LNCGme00344) with a novel locus (LNCGme00345), all of which are eRNAs and are bound by PAX5, differentially expressed in the absence of PAX5, and lie downstream of the B-cell lymphoma 11a gene (Bcl11a), whose expression is also PAX5-dependent in B-ALL cells (Figure 5C). We note that Bcl11a does not have PAX5 binding sites near its TSS, and yet its expression is PAX5-dependent. Taking these observations together, we hypothesize that LNCGme00432, LNCGme00344, and LNCGme00345 may be transcribed from PAX5-induced enhancers that in turn regulate expression of Bcl11a and thus explain how expression of Bcl11a is indirectly PAX5-dependent. Because BCL11A has been proposed to activate expression of the Rag1 and Rag2 genes and hence to promote recombination of V, D, and J immunoglobulin genes,39 it may contribute to PAX5-dependent differentiation of B-ALL cells and subsequent regression of the leukemia. Furthermore, BCL11A is essential for B-cell development40,41 ; thus, these lncRNAs may also mediate the effect of PAX5 on expression of Bcl11a during normal B-cell development. Interestingly, LNCGme00344 and LNCGme00345 have human eRNA orthologs (BMThy_chr2_0943 and BMThy_chr2_0945; supplemental Table 6), suggesting that this transcriptional regulation may be conserved in human B cells. In total, we report 199 lncRNAs (including 73 eRNAs) that are bound by PAX5, show PAX5-dependent expression, and are neighboring a protein-coding gene that also shows PAX5-dependent expression (supplemental Table 7).
Identification of lncRNAs with PAX5-dependent expression in B-ALL cells. (A) Venn diagram depicting overlap between lncRNAs with sufficient read coverage to be included in this analysis (black), lncRNAs differentially expressed (DE) between B-ALL cells with and without doxycycline-induced Pax5 expression (red), and PAX5 transcription factor binding sites (TFBS) annotated in either pro-B cells or mature B cells (blue) that could not be associated with a protein-coding gene. A subset of lncRNAs is both DE and has PAX5 bound within the gene body or promoter region (peach). (B) Volcano plot depicting the fold-change in lncRNA expression between B-ALL cells vs B-ALL cells with doxycycline-induced Pax5 expression (see supplemental Table 7) plotted against the probability that this difference had occurred by chance (q value). Each dot represents a single lncRNA and is colored black unless it was DE (q < .05) and either near or not near a PAX5 binding site (peach or red, respectively). (C) Genome plot showing PAX5-bound eRNA loci (LNCGme00432, LNCGme00344, and LNCGme00345), together with their proximal protein-coding gene, the zinc finger protein gene B-cell lymphoma 11a (Bcl11a). All are DE in B-ALL cells upon induction of Pax5 expression. PAX5 binding sites in pro-B cells are indicated in peach. The other annotated lncRNA (LNCGme00346) is also an eRNA that is DE on induction of Pax5 expression, but it shows no evidence of PAX5 binding.
Identification of lncRNAs with PAX5-dependent expression in B-ALL cells. (A) Venn diagram depicting overlap between lncRNAs with sufficient read coverage to be included in this analysis (black), lncRNAs differentially expressed (DE) between B-ALL cells with and without doxycycline-induced Pax5 expression (red), and PAX5 transcription factor binding sites (TFBS) annotated in either pro-B cells or mature B cells (blue) that could not be associated with a protein-coding gene. A subset of lncRNAs is both DE and has PAX5 bound within the gene body or promoter region (peach). (B) Volcano plot depicting the fold-change in lncRNA expression between B-ALL cells vs B-ALL cells with doxycycline-induced Pax5 expression (see supplemental Table 7) plotted against the probability that this difference had occurred by chance (q value). Each dot represents a single lncRNA and is colored black unless it was DE (q < .05) and either near or not near a PAX5 binding site (peach or red, respectively). (C) Genome plot showing PAX5-bound eRNA loci (LNCGme00432, LNCGme00344, and LNCGme00345), together with their proximal protein-coding gene, the zinc finger protein gene B-cell lymphoma 11a (Bcl11a). All are DE in B-ALL cells upon induction of Pax5 expression. PAX5 binding sites in pro-B cells are indicated in peach. The other annotated lncRNA (LNCGme00346) is also an eRNA that is DE on induction of Pax5 expression, but it shows no evidence of PAX5 binding.
Discussion
To aid in the identification of lncRNAs active within the immune system, we present a comprehensive catalog of 4516 lncRNAs, including 2349 intergenic loci, expressed across 11 murine B-cell populations. Translating genome-scale lncRNA identification into in vivo functional studies remains a major challenge that necessitates the use of model organisms. Previous work has identified lncRNAs expressed in a subset of these populations in humans17,33-35 ; however, given the high tissue specificity and rapid evolution of lncRNAs, a murine catalog is therefore a prerequisite to successful lncRNA studies in the mouse. In addition, we defined patterns of expression, chromatin modification, and transcription factor binding that provide insight into the regulation and function of these transcripts and we identified a subset of lncRNAs that have human orthologs and thus are of direct relevance to human research.
The majority (67%) of lncRNA loci present in our catalog contained no evidence of splicing. Although single-exon transcripts had to be replicated in 2 or more samples to be included in this catalog, their loci were less likely to be in published datasets. However, the high rate of polymerase chain reaction validation of these single-exon lncRNAs (85%) and their comparable intersection with clearly defined chromatin peaks confirms the reliability of the transcript annotation, and the demonstrated functions of single-exon lncRNAs such as NEAT142 and Paupar43 highlight the importance of including these transcripts in the catalog.
LncRNA genomic locations have the potential to provide insight into function. A substantial proportion of multiexon (48%) and single-exon (48%) loci were antisense to or flanking (<5 kb) protein-coding genes. Previous studies have classified as many as 60% of detected lncRNAs as antisense transcripts arising from bidirectional transcription at protein-coding promoters44 that could regulate expression of genes encoding transcriptional regulators.45 These are included within the lncRNAs classified as flanking in this study. Full-length antisense lncRNAs have also been implicated in a variety of cellular functions,46,47 most notably FAS-AS1, which regulates alternative splicing in its antisense coding gene Fas.5,48
Half of all lncRNAs detected in B cells (52%) were intergenic (>5 kb from a protein-coding gene). The use of H3K4me1 and H3K4me3 chromatin marks to distinguish eRNAs from pRNAs has been previously demonstrated,8 and we were able to corroborate many of the features of these transcripts. The consistent classification of these loci across B-cell development strongly suggests that these markers identify 2 distinct and mutually exclusive classes of lncRNA.
Comparison with lncRNAs expressed in human B cells identified 185 mouse lncRNAs that had a human ortholog and a further 228 mouse eRNAs that have human eRNA counterparts. This offers the possibility of using the power of mouse genetics to study the in vivo function of these human lncRNAs by genetic modification of the corresponding mouse genes.
The use of expression correlation as a means of associating lncRNA function with that of protein-coding genes has been successfully demonstrated in previous studies.17,18 By clustering protein-coding genes on the basis of coexpression, we were able to identify discrete groups whose patterns of expression transcend individual B-cell populations and whose members are associated with, for example, cell-cycle function. Post hoc association of lncRNAs with these clusters implicated a substantial proportion (22%) of the lncRNAs with these specific processes. In addition, this analysis supported the hypothesis that eRNAs may function to regulate expression of adjacent protein-coding genes.7
Using external data, we identified loci regulated by PAX5, a crucial mediator of B-cell development, whose loss contributes critically to the development of B-ALL.38 After incorporating our classification of lncRNAs based on chromatin state, we identified 73 eRNAs situated proximal to protein-coding genes that also show Pax5-dependent expression in B-ALL cells. The roles of such transcripts remain to be investigated; however, their discovery demonstrates the utility of our catalog when used in conjunction with external datasets to probe the complexities of B-cell development and disease.
In conclusion, RNA-seq followed by de novo transcript assembly has the potential to identify thousands of novel long noncoding transcripts. Although a growing number of these transcripts have been assigned to diverse cellular processes, as yet, the majority have no known function. Given the large numbers of these transcripts, a major challenge when seeking to identify and characterize lncRNA function lies in the prioritization of plausible candidates for in vivo functional studies. We anticipate that this catalog will serve as a valuable resource for such future research—for example, by allowing systematic genetic screens of high-confidence lncRNAs to identify those with key functions in B-cell development and activation.
The data reported in this article have been deposited at National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE72019).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Stephen Nutt for Blimp1-GFP mice.
This work was supported by the Flow Cytometry and Advanced Sequencing Facilities of the Medical Research Council (MRC) National Institute for Medical Research (now the Francis Crick Institute). The MRC Computational Genomics Analysis and Training Centre and V.L.J.T. were supported by the Medical Research Council (MRC program number U117527252). V.L.J.T. was supported by the Francis Crick Institute, which receives its funding from the MRC, Cancer Research UK, and the Wellcome Trust.
Authorship
Contribution: T.F.B., J.S.J., and J.M. performed research; T.F.B., J.S.J., J.M., and A.H. analyzed data; and T.F.B., J.S.J., A.H., C.P.P., and V.L.J.T. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Victor L. J. Tybulewicz, The Francis Crick Institute, The Ridgeway, London NW7 1AA, United Kingdom; e-mail: victor.t@crick.ac.uk.
References
Author notes
T.F.B. and J.S.J. contributed equally to this work