Key Points
Ribosome profiling of primary human platelets defines the platelet translatome, derived from a biased subset of MK mRNAs.
Restoration of the ribosome rescue/mRNA surveillance factor Pelota, which is normally absent in wild-type platelets, promotes RNA decay.
Abstract
Platelets are anucleate cytoplasmic fragments that lack genomic DNA, but continue to synthesize protein using a pool of messenger RNAs (mRNAs), ribosomes, and regulatory small RNAs inherited from the precursor megakaryocyte (MK). The regulatory processes that shape the platelet transcriptome and the full scope of platelet translation have remained elusive. Using RNA sequencing (RNA-Seq) and ribosome profiling of primary human platelets, we show the platelet transcriptome encompasses a subset of transcripts detected by RNA-Seq analysis of in vitro–derived MK cells and that these platelet-enriched transcripts are broadly occupied by ribosomes. We use RNA-Seq of synchronized populations of in vitro–derived platelet-like particles to show that mRNA decay strongly shapes the nascent platelet transcriptome. Our data suggest that the decay of platelet mRNAs is slowed by the natural loss of the mRNA surveillance and ribosome rescue factor Pelota.
Introduction
Platelets are small, highly abundant cell fragments that circulate in the blood and maintain hemostasis. They activate an array of clot-forming functions, including morphological changes, adhesion, and secretion, when they encounter molecular markers of injury. Nascent platelets form during cytoplasmic maturation of bone marrow megakaryocytes (MKs) and are released to the circulation in vast numbers each day.1,2 Newly released (“reticulated”) platelets retain MK-derived RNA3 ; this reticulated subpopulation is most evident when the rate of thrombopoiesis is elevated, which occurs after acute blood loss4 or during thrombocytopenia.5-11 Because platelets lack the ability to synthesize new messenger RNA (mRNA) transcripts or assemble new ribosomes, the abundance of MK-derived RNA in platelets gradually declines over several days.10,12,13 This bulk decrease in RNA principally reflects the clearance of ribosomes (and the constituent ribosomal RNA [rRNA]), similar to the maturation process that occurs in reticulocytes during terminal differentiation.14-16 The mechanisms of mRNA and ribosome decay in platelets are not known. Autophagy-dependent ribosome degradation processes have been identified in yeast17 as well as in reticulocytes,14 where defective ribosome or organelle clearance is associated with mild anemia and, in mice, an increased hemoglobin content.14 Interestingly, however, the ribosome-associated endonuclease SLFN14 (which is presumably distinct from the autophagy-related machinery) has been proposed to be involved in rRNA degradation in reticulocytes,18 and was recently implicated in functional platelet deficiencies and thrombocytopenia in humans.19,20 These observations suggest that platelets use a different ribosome clearance mechanism than reticulocytes.
Using next-generation RNA sequencing (RNA-Seq), the work of several groups has greatly expanded our knowledge21,22 of the platelet transcriptome, confirming that its complexity is comparable to a typical mammalian cell23-27 and is shaped by regulatory processes such as mRNA sorting by MKs28 and regulation by microRNAs.29 As with mRNAs in other tissues, platelet mRNAs are marked by 5′ m7G-caps30 and 3′polyA-tails,25,31 which are important features recognized by the eukaryotic translational machinery. Numerous specific examples of proteins synthesized by platelets have been described,32-36 and an active microRNA pathway has been studied26,27,29,37,38 and implicated in regulating mRNA decay and translation in platelets.29
Because it is technically difficult to study platelet populations of different ages in vivo,39 RNA-Seq studies have reported on an “ensemble” average of platelet transcripts that are influenced by the underlying age distribution of circulating platelets at the time of sample collection. Given the documented clearance of RNA by platelets during the first several days of circulation,3,10,12 it seems likely that platelet protein synthesis primarily occurs within a few days of release from the MK.11,13 However, some evidence exists for translation in older platelets.40 This time course is comparable to that of protein synthesis and subsequent translational arrest during reticulocyte maturation,41,42 which has been proposed to be caused by either clearance of ribosomes14 or the accumulation of inactive ribosomes.43
Here, we combine ribosome profiling with mRNA sequencing to survey active gene expression in primary platelets. We find that platelets inherit a subset of MK mRNAs and translate these transcripts, although published mass spectrometry data show the accumulation of protein products is biased toward those with a function in platelets and away from, for example, nuclear components. Our data suggest thrombin activation increases bulk protein synthesis and leads to mRNA-specific translational activation and repression. We model platelet production in culture and find prominent roles of both mRNA and rRNA decay, pointing to the loss of gene expression as a molecular feature of platelet maturation and aging. Finally, we show that RNA decay in platelets is modulated by the ribosome rescue and quality control factor Pelota (PELO), which we have shown plays a specialized role in both platelet and reticulocyte translation.
Methods
Platelet preparations
Platelets were obtained from 3 healthy volunteers by single-donor apheresis followed by additional leukoreduction to achieve <1 × 106 white blood cells per unit of >1 × 1011 platelets. Apheresis samples were collected by HemaCare (Van Nuys, CA) in accordance with institutional review board #00001463 (Johns Hopkins University). Platelets were stored on a rotating platform at room temperature for <2 hours and used within 26 hours of apheresis.
Platelet ribosome profiling
Ribosome profiling is a high-throughput strategy that relies on the deep sequencing of ribosome protected fragments of mRNA.44 This technique first involves cell lysis under conditions that stabilize native interactions between ribosomes and mRNAs.45 Next, unprotected mRNA is digested with a purified ribonuclease (Escherichia coli RNase I); the resulting short, ∼28 to 32 nucleotide fragments protected by the ribosome are converted to DNA and analyzed by deep sequencing. Alignment of short sequencing reads back to the reference genome reports on the specific positions of ribosomes along mRNAs in the cell.46 For platelets, these steps were carried out essentially as previously described,45 but with the following modifications. Platelets were counted manually using a hemocytometer and ∼1 × 108 platelets were used per ribosome profiling experiment from each of a total of 3 healthy human volunteers. Platelets were released from the apheresis unit in tubes containing the oligo peptide Gly-Pro-Arg-Pro47,48 to block fibrin polymerization and fibrinogen, as described,36 before lysis in polysome lysis buffer.45
RNA-Seq
For RNA-Seq experiments, total platelet RNA was prepared from ∼1 × 108 platelets by direct lysis in Trizol (Life Technologies) and ethanol precipitation using 1 mg/mL GlycoBlue (Ambion) as a carrier. Integrity of total RNA was analyzed using a RNA Pico 6000 BioAnalyzer Chip (Agilent). A total of 100 ng to 1 µg of total RNA per sample was used for RNA-Seq library preparation using the mRNA strand-specific TruSeq kit with RiboZero-Gold to deplete rRNAs (Illumina).
Meg/PLP cell culture
Meg01 cells were purchased from ATCC and grown in RPMI 1640 supplemented with 10% fetal bovine serum. Platelet-like particles (PLPs)49-52 were purified from Meg01 cell culture supernatant for 5 to 7 days. Cells were removed from supernatants by centrifugation at 100g × 5 minutes and then filtered using a 5-µm filter (Millipore). PLPs were pelleted at 3000g × 10 minutes and resuspended in 1X phosphate-buffered saline.
PLP aging
Meg01 cell cultures were grown in semisuspension culture until reaching a density of 1 × 106/mL. Activating anti-Fas antibody (Millipore clone CH11) was then added at 10 ng/mL and PLPs were purified from the media (as previously) after 24 hours, and purity and yield were determined by flow cytometry. Two replicates of purified PLPs collected over 24 hours were then aged for each time point in standard cell culture conditions.
Platelet activation experiments
A total of 1 × 108 platelets from leukodepleted apheresis samples were washed in Tyrode albumin buffer (NaCl, 136.5 mM; KCl, 2.6 mM; NaHCO3, 11.9 mM; Na2H2PO4, 0.43 mM; MgCl2, 1 mM; CaCl2, 2 mM; glucose, 5.5 mM; bovine serum albumin, 0.35%) and human thrombin (Sigma) was added to a final concentration of 0.01U/mL. The oligopeptide Gly-Pro-Arg-Pro47,48 was also added to these reactions to block fibrin polymerization, which prohibits adequate release of polysomes from platelets. Activation reactions were then incubated at 37°C on a nutator for 60 minutes before harvesting.
Gene ontology analyses
Ontology analyses were performed on ranked gene lists using the Gorilla Gene Ontology Program (http://cbl-gorilla.cs.technion.ac.il/).53 Enrichment of specific functional categories was determined using corrected P values according to a Benjamini and Hochberg false discovery rate.54
Differential expression analysis
Differential gene expression in RNA-Seq or ribosome profiling data were analyzed using DESeq2 (BioConductor) with standard parameters.55,56 For time series analysis, we performed likelihood ratio tests using an overall model (genotype + time + genotype:time) and a reduced model (genotype + time).57 Processing of ribosome profiling data was performed as described previously.45
Published datasets
Integrated platelet mass spectrometry data used to estimate protein copy numbers based on spectral counts58,59 were downloaded from the Pax database (http://pax-db.org/). Ensembl protein identifiers were matched to University of California Santa Cruz identifiers using the BioMart (http://www.biomart.org/) function in R. Platelet mass spectrometry data from Burkhart et al60 were downloaded from the Proteomics Identifications archive (http://www.ebi.ac.uk/pride/; accession numbers 22201-22203, 22206). Human ex vivo differentiated MK RNA-Seq61 data were downloaded as bam files from European Molecular Biology Laboratory-European Bioinformatics Institute ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under the experiment identifier E-MTAB-918.
Results
Platelet mRNAs are broadly occupied by ribosomes
We analyzed translation in primary human platelets using ribosome profiling and high-throughput RNA-Seq following rRNA depletion.45 As expected, libraries prepared by both methodologies were enriched in sequencing reads derived predominantly from mRNAs (Figure 1A). Given their purely cytoplasmic nature, we observed far fewer intronic reads in platelets as well (<1% vs 10-20% in typical nucleated cells62,63 ). Ribosome profiling libraries contained relatively more transfer RNA fragments and fewer reads mapping to long, intergenic, noncoding RNAs when compared with the RNA-Seq libraries. Ribosome profiling identified >6000 different ribosome-occupied mRNAs in platelets using a reads per kilobase per million mapped (RPKM) threshold of 0.3. Gene ontology analysis53 of these transcripts identified significant enrichment for processes associated with known platelet functions such as chemokine/cytokine signaling and platelet activation/degranulation (Figure 1B). This suggests platelets play an active role in synthesizing proteins crucial to principal platelet functions. Concordance was high between platelet RNA-Seq and ribosome profiling data (Spearman ρ = 0.77) and the highest expressed genes, TMSB4X, B2M, PPBP, F13A1, PF4, CCL5, NRGN, and SDPR, in both data sets encode well-known platelet proteins (Figure 1C-D).23
Ribosome profiling of human platelets. (A) Fraction of sequencing reads from RNA-Seq and ribosome profiling libraries mapped to genomic features. lincRNA, long, intergenic, noncoding RNA; tRNA, transfer RNA. (B) Gene ontology analysis of translated mRNAs (single ranked list; see “Methods”). (C) Numbers of unique transcripts identified by ribosome profiling (Ribo-Prof) and RNA-Seq. Yellow box indicates the sensitivity threshold 0.3 RPKM. (D) Scatter plot showing the correlation between RNA-Seq and ribosome profiling data from platelets (Spearman ρ = 0.77). (E,F) Analysis of the effects of 0.01 U/mL human thrombin for 60 minutes at 37°C on platelet translation. Blue, mitochondrial transcripts; FP, footprints; pos, positive; red, significant changes (adjusted P < .05); reg, regulation.
Ribosome profiling of human platelets. (A) Fraction of sequencing reads from RNA-Seq and ribosome profiling libraries mapped to genomic features. lincRNA, long, intergenic, noncoding RNA; tRNA, transfer RNA. (B) Gene ontology analysis of translated mRNAs (single ranked list; see “Methods”). (C) Numbers of unique transcripts identified by ribosome profiling (Ribo-Prof) and RNA-Seq. Yellow box indicates the sensitivity threshold 0.3 RPKM. (D) Scatter plot showing the correlation between RNA-Seq and ribosome profiling data from platelets (Spearman ρ = 0.77). (E,F) Analysis of the effects of 0.01 U/mL human thrombin for 60 minutes at 37°C on platelet translation. Blue, mitochondrial transcripts; FP, footprints; pos, positive; red, significant changes (adjusted P < .05); reg, regulation.
Platelet activation with thrombin has previously been associated with an increase in bulk platelet protein synthesis through an mTOR-dependent mechanism.34,36 To comprehensively test how platelet activation affects translation of specific mRNAs, we performed ribosome profiling on platelets activated with thrombin for 60 minutes at 37°C. To estimate global protein synthesis, we quantified the increase in overall cytosolic ribosome footprints after normalization to footprints derived from mitochondrial ribosomes,64,65 which are entirely distinct from the cytosolic translational machinery and are continuously synthesized in platelets.66 These analyses show there is a global increase in ribosome occupancy on cytoplasmic mRNAs, consistent with increased translational output (Figure 1E-F; gray vs blue transcripts). This effect may reflect increased initiation of translation by previously free ribosomes, but also decreased termination and subunit dissociation. We also cannot exclude the possibility that activation by thrombin leads to a global decrease in mitochondrial translation. Furthermore, the translation of certain mRNAs was significantly more affected than others (Figure 1E-F; red transcripts, P < .05). For example, ribosome occupancy on F13A1, PFN1, and MYL6 transcripts increased after thrombin activation (1.5- to twofold; P < .05), whereas ribosome occupancy of CLL5, PPBP, and ANGPT1 decreased (1.6- to 2.5-fold; P < .05). These changes could reflect the immediate demand for structural proteins involved in hemostatic plug formation and influence later immune functions of cytokine and chemokine release by activated platelets.67
Platelet transcriptome, translatome, and proteome correlations
For a fully integrated view of platelet gene expression, we compared our platelet RNA-Seq and ribosome profiling data to available protein abundance data from mass spectrometry.58-60,68 We included 8801 platelet transcripts and 6809 translated mRNAs from our data along with 4147 quantified proteins (Figure 2A). Within the 2246 genes that were supported by all 3 data types, we observed a moderate, statistically significant correlation between either platelet mRNA expression level or ribosome footprint density and protein abundance (Spearman ρ = 0.39 and ρ = 0.37, respectively) (Figure 2B; supplemental Figure 1A, available on the Blood Web site). As anticipated, ribosome density was slightly higher on transcripts detected by mass spectrometry (supplemental Figure 1B), consistent with detectability bias favoring abundant proteins.
Integrated analysis of the platelet transcriptome, translatome, and proteome. (A) Number of platelet transcripts identified by RNA-Seq (this study), ribosome profiling (this study), or mass spectrometry (mass-spec; published datasets). The degree of overlap is to scale and major gene ontology (GO) categories are listed (and listed comprehensively in supplemental Tables 1-8). (B) Scatter plot showing the correlation between ribosome profiling and protein abundance in platelets (Spearman ρ = 0.37). (C) GO analysis of the 2246 high-confidence genes detected by RNA-Seq, ribosome profiling, and mass spectrometry.
Integrated analysis of the platelet transcriptome, translatome, and proteome. (A) Number of platelet transcripts identified by RNA-Seq (this study), ribosome profiling (this study), or mass spectrometry (mass-spec; published datasets). The degree of overlap is to scale and major gene ontology (GO) categories are listed (and listed comprehensively in supplemental Tables 1-8). (B) Scatter plot showing the correlation between ribosome profiling and protein abundance in platelets (Spearman ρ = 0.37). (C) GO analysis of the 2246 high-confidence genes detected by RNA-Seq, ribosome profiling, and mass spectrometry.
Prior studies have underscored greater than expected discordance between measurements of platelet transcripts by high-throughput sequencing and proteomics-based measurements of protein levels.61,69-71 We next sought to better understand this discordance using our ribosome profiling data and determine the extent to which it may reflect biological phenomena such as uptake/sequestration of serum proteins,72 or other unusual features of platelet gene expression,13 vs technical artifacts of the different methods used to measure nucleic acids and proteins.73,74 High-confidence genes identified by all 3 methods (RNA-Seq, ribosome profiling, and mass spectrometry), which were both synthesized and accumulated in platelets, were significantly enriched for cytoplasmic processes such as “intracellular transport” and “gene expression” (q value < 0.05) (Figure 2C) and depleted for nuclear functions, consistent with the lack of nuclei in platelets (supplemental Tables 1-3). In contrast, genes that were detected by RNA-Seq and ribosome profiling, but not by mass spectrometry, were enriched in nuclear components and functions such as transcription and chromatin organization (supplemental Figure 2A; supplemental Tables 4-6), perhaps suggesting rapid degradation of unneeded nuclear proteins in platelets. Genes whose products were detected by mass spectrometry only and no other method were strongly enriched in skin and serum proteins (supplemental Figure 2B; supplemental Tables 7 and 8) and likely reflect contaminants or proteins taken up from serum rather than expressed in platelets.72 These 3 orthogonal approaches define a conservative, but high-confidence view of the platelet transcriptome, translatome, and proteome.
Platelets contain a subset of MK mRNAs
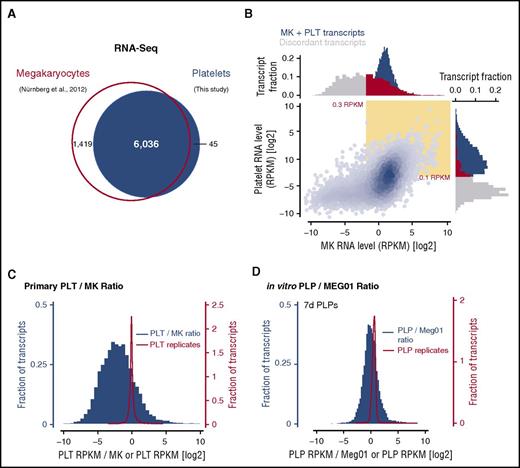
Concordance between our platelet transcriptome and published RNA-Seq data from human MKs derived by in vitro differentiation of cord blood CD34+ hematopoietic progenitor cells61 was high, with 6036 (∼80%) of transcripts shared between MKs and platelets, and transcripts that were discordant between the samples (∼20%) were almost exclusively MK-specific (Figure 3A). Whereas approximately half of the MK-specific transcripts were expressed at high levels (Figure 3B, top gray marginal histogram), transcripts detected in platelets only were almost uniformly of low abundance (Figure 3B, side gray marginal histogram), below the established sensitivity threshold for detection. These results are consistent with the expectation that platelet mRNAs encompass a subset of transcripts synthesized in MKs, and MK-specific transcripts likely represent either mRNAs preferentially retained by MKs (rather than packaged into nascent platelets), or transcripts that are rapidly degraded by young platelets.
Comparative analysis of the platelet (PLT) and megakaryocyte transcriptomes. (A) Number of unique transcripts identified by RNA-Seq in megakaryocytes (Nürnberg et al, 2012)61 and platelets (this study). (B) Transcript abundance in megakaryocytes and platelets based on RNA-Seq data. Yellow inset indicates transcripts expressed above the determined threshold level (0.3 or 0.1 RPKM). Marginal histograms show the expression of discordant transcripts in megakaryocytes and platelets (gray and red) or shared transcripts (blue). (C) Histograms of platelet enrichment scores (platelet RPKM/MK RPKM; blue) or RNA-Seq replicates (platelet replicate 1 RPKM/platelet replicate 2 RPKM; red). (D) Identical analysis as in panel C performed for in vitro–derived PLPs and corresponding Meg01 samples.
Comparative analysis of the platelet (PLT) and megakaryocyte transcriptomes. (A) Number of unique transcripts identified by RNA-Seq in megakaryocytes (Nürnberg et al, 2012)61 and platelets (this study). (B) Transcript abundance in megakaryocytes and platelets based on RNA-Seq data. Yellow inset indicates transcripts expressed above the determined threshold level (0.3 or 0.1 RPKM). Marginal histograms show the expression of discordant transcripts in megakaryocytes and platelets (gray and red) or shared transcripts (blue). (C) Histograms of platelet enrichment scores (platelet RPKM/MK RPKM; blue) or RNA-Seq replicates (platelet replicate 1 RPKM/platelet replicate 2 RPKM; red). (D) Identical analysis as in panel C performed for in vitro–derived PLPs and corresponding Meg01 samples.
Even among the transcripts present in both platelets and MKs, we found broad quantitative variance in mRNA abundance (Figure 3B, blue scatter plot; Figure 3C). The abundance ratio between platelets and MKs covered more than 3 standard deviations from the mean (supplemental Figure 3A). Similar variation was observed using published platelet RNA-Seq datasets from Rowley et al (supplemental Figure 3B).23 Again, these data point to an underlying active process such as MK mRNA sorting28 or platelet mRNA decay11,13 shaping the platelet transcriptome and creating the broad variation in platelet enrichment we observed. A limitation of this analysis, however, is that the cultures of ex vivo–differentiated MKs used for RNA-Seq by Nürnberg et al contained 0% to 10% undifferentiated cells.61 The extent to which the RNAs contained in this fraction of contaminating cells contributes to this comparison, or the extent to which the transcriptome of ex vivo–differentiated MKs differ from the in vivo MK transcriptome, is unknown.
To address this uncertainty and further explore the mechanisms that sculpt the platelet transcriptome from the population of MK mRNAs, we took advantage of an in vitro PLP-generating system49-51 to isolate synchronized populations of PLPs at specific times after release by Meg01 cells. Importantly, PLPs derived from Meg01 cells are structurally and functionally similar to primary platelets.49-52 We first prepared RNA-Seq libraries from matched Meg01 cells and PLPs harvested over 7 days ( to approximate the age distribution of primary human platelets in vivo). We observed strong overlap between Meg01 and PLP transcriptomes (supplemental Figure 3C) and, using these data, we calculated enrichment scores for the abundance of PLP transcripts relative to Meg01 cells. This analysis revealed a strikingly broad range of enrichment (Figure 3D; supplemental Figure 3C), similar to the mRNA abundance differences we observed between primary human platelets and ex vivo MKs. The enrichment distribution was also left-skewed, reflecting that the PLP transcriptome is derived from the Meg01 transcriptome by either (1) retention/sorting of specific mRNAs in the MK or (2) degradation of certain RNAs by platelets.
RNA decay helps shape the nascent PLP transcriptome
To explore these 2 mechanisms, we next sought to characterize changes in the platelet transcriptome occurring shortly after their release. We analyzed synchronized populations of newly released PLPs by high-throughput RNA-Seq during a time course of 0 to 6 hours (Figure 4A). Because cytoplasmic RNA levels are not at steady state in PLPs, we used the levels of mitochondrial transcripts to normalize our expression measurements and further validated this strategy by adding exogenous control RNA to each sample (Figure 4B-C). We found that cytoplasmic mRNA abundance declined substantially beginning at 4 hours after PLP collection as the levels of mitochondrial transcripts began to compose a substantially larger fraction of total RNA (Figure 4D-F). Based on these data, the typical mRNA decays with a half-life of 5.7 hours in PLPs (supplemental Figure 4A), consistent with recent measurements of bulk RNA turnover in primary platelets.11 Histone mRNAs were somewhat shorter-lived (average half-life, 4.2 hours), whereas ribosomal protein mRNAs were slightly longer-lived than average (average half-life, 6.6 hours) (supplemental Figure 4B-C), and a few specific mRNAs seemed to be especially resistant to degradation, including MYD88 and RAP1A, which have special roles in platelets75-78 (Figure 4D). Taken together, these data suggest that RNA decay is likely a major driver that determines transcript abundance (and therefore to a significant extent translation) in platelets.
Global analysis of mRNA decay by PLPs. (A) Experimental design for analysis of mRNA decay by PLPs. (B) BioAnalyzer pseudo gel showing degradation of 28 and 18S cytoplasmic rRNAs during PLP aging between 1 and 2 days after purification (see “Methods”). (C) Quantification of 28S (blue diamonds) and 18S (gray diamonds) degradation in (B) compared with quantitative reverse transcription polymerase chain reaction of mitochondrial (mito) 16S rRNA (black diamonds). (D) M-A plot showing changes in transcript abundance between 4 and 6 hours and 1 and 2 hours in PLPs. Histone category includes replication-type nonpolyadenylated mRNAs and replication-independent polyadenylated histone mRNAs. (E,F) Plots of average RNA abundance for gene groups over a 0- to 6-hour time course. FLUC, firefly luciferase; M-A, ratio-average; RP, ribosomal protein.
Global analysis of mRNA decay by PLPs. (A) Experimental design for analysis of mRNA decay by PLPs. (B) BioAnalyzer pseudo gel showing degradation of 28 and 18S cytoplasmic rRNAs during PLP aging between 1 and 2 days after purification (see “Methods”). (C) Quantification of 28S (blue diamonds) and 18S (gray diamonds) degradation in (B) compared with quantitative reverse transcription polymerase chain reaction of mitochondrial (mito) 16S rRNA (black diamonds). (D) M-A plot showing changes in transcript abundance between 4 and 6 hours and 1 and 2 hours in PLPs. Histone category includes replication-type nonpolyadenylated mRNAs and replication-independent polyadenylated histone mRNAs. (E,F) Plots of average RNA abundance for gene groups over a 0- to 6-hour time course. FLUC, firefly luciferase; M-A, ratio-average; RP, ribosomal protein.
Decline of PELO levels slows mRNA decay in PLPs
In light of these data, we were interested in understanding the molecular mechanisms that result in the degradation of certain mRNAs in young platelets. We recently described dynamic regulation of ribosome recycling and rescue factors in anucleate blood lineages, including platelets and reticulocytes.79 Both lineages lose ABCE1, the canonical ribosome recycling factor required after translation termination. Ultimately, both also lose PELO, a ribosome rescue/mRNA surveillance factor required for the release of stalled ribosomes from truncated mRNAs and that plays a role in promoting mRNA decay.80-82 MKs express both PELO and ABCE1, yet platelets (and Meg01-derived PLPs) naturally lack PELO and contain only low amounts of ABCE1. We previously reported that reduced levels of ABCE1 and PELO result in the accumulation of unrecycled ribosomes in the 3′UTRs of mRNAs, limiting the availability of ribosomes.79 Given the role of RNA decay in platelets, we hypothesized that the naturally reduced levels of ribosome rescue/recycling in platelets may play a role in slowing mRNA decay in platelets, thereby preserving mRNAs and promoting translation.
We thus tested whether restoration of PELO in PLPs impacts the rates of mRNA decay. We first prepared Meg01 cell lines that transgenically overexpress the PELO protein and characterized the changing transcriptome of derived PLPs over the same 6-hour time course described previously (Figure 4A). The initial transcriptomes of PELO- and GFP-overexpressing PLPs were highly similar (Figure 5A). The PELO mRNA was overexpressed ∼12-fold; as expected, this increase in PELO mRNA was associated with a significant increase in levels of HBS1L transcript (which encodes the PELO cofactor) as well as increased hemoglobin γ 1/2 transcript levels (Figure 5A-B). To account for changes in transcript abundance that occur during the time course in both GFP and PELO samples, we incorporated PLP age and Meg01 genotype as independent factors in a generalized linear model, with PELO-dependent changes to mRNA decay emerging as interactions between these 2 factors. Histone transcript decay was further accelerated by PELO overexpression (average half-life, 3.0 hours compared with 4.2 hours in GFP control PLPs; Figure 5C; supplemental Figure 4C), consistent with the known role for PELO in targeting histone mRNAs for decay in yeast.83 Interestingly, transgenic overexpression of PELO resulted in more rapid degradation of most cytoplasmic PLP transcripts, as visualized by the relative reduction in cytoplasmic transcript abundance vs mitochondrial transcripts (average half-life, 3.7 vs 5.7 hours in control PLPs; Figures 5C and 4A). Importantly, our analysis is not sensitive to differences in initial transcript abundance or to changes that occur over time in both conditions (Figure 5D); instead, our analysis tests for the presence of specific effects of PELO overexpression on transcript abundance changes. Taken together, these data show that PELO accelerates the degradation of cytoplasmic PLP transcripts and suggest that the natural absence of PELO from platelets may function to globally slow mRNA decay and thereby preserve mRNAs. In the absence of ongoing RNA synthesis, such reduced mRNA decay may be necessary for abundant protein synthesis in platelets.
Natural loss of PELO slows mRNA decay in PLPs. (A) M-A plot showing changes in transcript abundance at the initial time point (“hour 0”) between PLPs overexpressing PELO vs a GFP control. (B) Rescaled window of the plot in panel A showing baseline differences in transcript abundance in PELO-overexpressing PLPs. (C) M-A plot showing the results of multivariate analysis to define changes in transcript abundance resulting from PELO overexpression after 1 to 2 hours (see “Methods”). (D) Heat maps depicting all log2FoldChanges for the 1000 transcripts with the lowest adjusted P values separating time (1-2 vs 4-6 hours) and condition (PELO vs GFP) variables. (E) Model showing differential sorting of mRNA transcripts to nascent platelets (left) and regulated mRNA decay of platelet mRNAs (right).
Natural loss of PELO slows mRNA decay in PLPs. (A) M-A plot showing changes in transcript abundance at the initial time point (“hour 0”) between PLPs overexpressing PELO vs a GFP control. (B) Rescaled window of the plot in panel A showing baseline differences in transcript abundance in PELO-overexpressing PLPs. (C) M-A plot showing the results of multivariate analysis to define changes in transcript abundance resulting from PELO overexpression after 1 to 2 hours (see “Methods”). (D) Heat maps depicting all log2FoldChanges for the 1000 transcripts with the lowest adjusted P values separating time (1-2 vs 4-6 hours) and condition (PELO vs GFP) variables. (E) Model showing differential sorting of mRNA transcripts to nascent platelets (left) and regulated mRNA decay of platelet mRNAs (right).
Discussion
The RNA content of platelets was recently discovered to contain unanticipated complexity,21,23-27,37 and the platelet transcriptome has been associated with important functional consequences.29,38,84-86 Through ribosome profiling, we show that much of this transcriptional complexity indeed shapes the platelet proteome through protein synthesis by platelet ribosomes. We combine our high-throughput RNA-Seq and ribosome profiling data with published mass spectrometry studies to present the first integrated view of platelet gene expression. Our analyses reveal important insights into the dynamics and underlying processes shaping the transcriptome, translatome, and proteome of platelets.
Our comparative analyses of MK and platelet transcriptomes suggest that a selective process, and not random sampling, dictates the content of MK-derived RNAs in platelets or PLPs. To explain the broad range of transcript enrichments we calculated in both MKs/platelets and Meg01/PLPs, we discuss several processes as potential mechanisms for this range, including specific sorting/retention of transcripts by MKs or regulated mRNA decay in nascent platelets (Figure 5E). Our analysis of mRNA sorting in Meg01/PLPs indicates the most well-sorted transcripts may be enriched/depleted as much as 16- to 32-fold (Figure 3D), which is approximately comparable to our analyses of the average effect of mRNA degradation, which shows a twofold reduction in bulk mRNA levels over 6 hours, that when compounded over the first 24 to 48 hours after platelet release, leads to a similar approximately 16- to 32-fold effect size (Figure 4D-F).
Independent support for MK RNA sorting28 and regulated mRNA decay26,29 in platelets has been described recently. Specifically, our results provide additional validation for the differential sorting of TIMP1-3, as well as MMP-1, MMP-2, and MMP-14 reported by Cecchetti et al.28 A recent study by Alhasan et al reported a substantial enrichment of circular RNA species in anucleate cell types relative to nucleated cells (including MKs) and attributed this effect to the increased impact of decay in anucleate cells coupled with the relative decay resistance of circular transcripts.13 Our study, as with that of Alhasan et al, suggests that RNA decay processes play an important role in shaping the platelet transcriptome. In our analysis, transcripts reported to give rise to circular RNA species behaved qualitatively similar to those that do not, although we do not address either the abundance or the degradation of circular RNAs directly (supplemental Figure 6).
A key prediction of both RNA decay and RNA sorting models is the existence of a population of transcripts that is present in MKs but depleted (or absent) in platelets, a phenomenon which is clearly evident in our data (Figure 3A-D). The extent to which these processes exhibit selectivity for platelet transcripts with certain specific functions warrants future investigation. Using an in vitro–derived platelet model, we directly tested the contribution of mRNA decay to transcript levels in young PLPs and found a role for the mRNA surveillance/ribosome rescue factor, PELO, in regulating global mRNA decay. We propose that overexpression of PELO releases unrecycled ribosomes from PLP 3′UTRs and stimulates mRNA degradation. A contributing factor for the slowed RNA decay in PELO-deficient (ie, wild-type) platelets could be sequestration by the ribosome of regulatory elements in the 3′ UTR of mRNAs that normally lead to exonucleolytic degradation.87,88
Finally, ongoing protein synthesis in platelets is subject to dynamic changes during normal aging or activation, and enriches the functional repertoire of these cell fragments. Our analysis provides the first comprehensive picture of translation in platelets, complementing work to define the transcriptome as well as the proteome, which reflects a mixture of inherited and newly synthesized proteins. It should serve as a valuable resource to understand normal platelet function and its disruption in disease.89-95
The data used in this article have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession number GSE94384).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by an American Heart Association predoctoral fellowship (E.W.M.), Medical Scientist Training Program funds (E.W.M.), Searle Scholars Program (Grant 11-SSP-229) (N.T.I.), and by the Howard Hughes Medical Institute (R.G.). This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California Berkeley, which is supported by the National Institutes of Health, National Center for Research Resources (S10 Instrumentation grants S10RR029668 and S10RR027303).
Authorship
Contribution: E.W.M. performed the experiments and analyzed data; R.G. and N.T.I. supervised the work and analyzed data. All authors wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rachel Green, Department of Molecular Biology and Genetics, Johns Hopkins University School of Medicine, 725 N Wolfe St, Baltimore, MD 21205; e-mail: ragreen@jhmi.edu; and Nicholas T. Ingolia, Department of Molecular Cell Biology, Center for RNA Systems Biology, Glenn Center for Aging Research, University of California Berkeley, 16 Barker Hall #3302, Berkeley, CA 94720; e-mail: ingolia@berkeley.edu.