Key Points
hEBI2 (GPR183) expression in mice leads to an abnormally expanded CD5+ B1a B-cell subset.
Mice expressing hEBI2 develop late-onset lymphomas similar to CLL.
Abstract
Human and mouse chronic lymphocytic leukemia (CLL) develops from CD5+ B cells that in mice and macaques are known to define the distinct B1a B-cell lineage. B1a cells are characterized by lack of germinal center (GC) development, and the B1a cell population is increased in mice with reduced GC formation. As a major mediator of follicular B-cell migration, the G protein–coupled receptor Epstein-Barr virus–induced gene 2 (EBI2 or GPR183) directs B-cell migration in the lymphoid follicles in response to its endogenous ligands, oxysterols. Thus, upregulation of EBI2 drives the B cells toward the extrafollicular area, whereas downregulation is essential for GC formation. We therefore speculated whether increased expression of EBI2 would lead to an expanded B1 cell subset and, ultimately, progression to CLL. Here, we demonstrate that B-cell–targeted expression of human EBI2 (hEBI2) in mice reduces GC-dependent immune responses, reduces total immunoglobulin M (IgM) and IgG levels, and leads to increased proliferation and upregulation of cellular oncogenes. Furthermore, hEBI2 overexpression leads to an abnormally expanded CD5+ B1a B-cell subset (present as early as 4 days after birth), late-onset lymphoid cancer development, and premature death. These findings are highly similar to those observed in CLL patients and identify EBI2 as a promoter of B-cell malignancies.
Introduction
Chronic lymphocytic leukemia (CLL) originates from clonal expansion of mature B cells expressing the B1 and T-cell marker CD5 and is the most common lymphoid cancer in humans.1 To understand the pathogenesis behind CLL, several mouse models mimicking the genetic aberrations observed in human CLL have been generated (see Simonetti et al2 for a review of current CLL mouse models). These models share the indolent disease course observed in human CLL, but moreover, the cancers seem to originate in B1a cells as indicated by the expression of CD5 and immunoglobulin M (IgM), low levels of CD23, and unmutated immunoglobulin heavy chain (IgH) variable genes.2 B1 cells secrete polyreactive and natural antibodies as part of the T-cell–independent humoral immune responses and, together with marginal zone (MZ) B cells, make up the innate-like B cells.3 B1 cells are predominantly found in the peritoneal and pleural cavities and only constitute 1% to 2% of the mature B-cell compartment in the spleen.4 Unlike B2 cells, which are produced continuously in the bone marrow, mouse B1 cells are generated only from hematopoietic stem cells in the fetal liver or in the bone marrow the first few weeks after birth and are subsequently maintained by self-renewal in the periphery.4
Recent studies have shown that downregulation of the G protein–coupled receptor (GPCR), Epstein-Barr virus (EBV)-induced gene 2 (EBI2 or GPR183), is important for participation in germinal center (GC) reactions, as knockout of the EBI2 gene facilitates B-cell migration to GCs, whereas retrovirus-induced expression retains the B cells in the extrafollicular area within secondary lymphoid organs.5,6 Thus, EBI2 functions as a chemotactic receptor responding to a finely tuned distribution of its ligand, oxysterol 7α,25-OH, which binds to EBI2 with high affinity and selectivity.7-11 Furthermore, previous studies have revealed that EBI2 induces cell proliferation12 and strong G protein–coupled signaling13-15 similar to the oncogenic GPCRs CMV-US28 and HHV8-ORF74,16-18 which may contribute to the maintenance of immune cells that express the receptor. To elucidate the role of EBI2 in B-cell and lymphoma development, we generated transgenic C57BL/6 mice expressing human EBI2 (hEBI2) under the control of the IgH promoter and intronic enhancer to induce expression in B cells (designated IgH-hEBI2). Here, we show that B-cell–targeted expression of hEBI2 in mice not only leads to an expanded CD5+ B1a B-cell subset from a young age, but also development of a late-onset CLL-like disease with lymphomatous transformation and premature death. In addition, the B2 cell compartment and, as a consequence, the GC-dependent humoral immune response are compromised.
Materials and methods
Generation of IgH-EBI2 and BCL-2xIgH-hEBI2 mice
The hEBI2 gene was cloned into an intronic IgH enhancer/promoter-driven vector and the sequence verified using an Alfexpress sequencer (Amersham Pharmacia Biotech, Piscataway, NJ). Generation of transgenic founders was done by pronuclear injection in CBAF1 hybrid blastocysts. Germ line transmission was confirmed by quantitative polymerase chain reaction (qPCR) amplification and Northern blot analysis from mouse tail DNA and splenic RNA, respectively. Transgenic animals were mated to C57BL/6, for >20 generations in the case of the original transgenic line. BCL-2 mice were originally developed by Harris and colleagues19 and obtained from The Jackson Laboratory. BCL-2xIgH-hEBI2 double transgenic mice were produced by crossing BCL-2 mice with IgH-hEBI2 mice. All genotypes were identified using primers listed in supplemental Table 1 (see supplemental Data, available on the Blood Web site). The use of mice in this study followed protocols approved by the veterinarian unit at the University of Copenhagen and the National Animal Experiments Inspectorate.
Real-time qPCR on tissue
RNA from snap-frozen tissue or fluorescence-activated cell sorting (FACS)-purified or magnetic-activated cell sorting (MACS)-purified cells was extracted using the Qiazol Lysis Reagent (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNA was digested by use of the TURBO DNA free kit (Ambion, Carlsbad, CA). Approximately 1 μg of total RNA was reverse transcribed with Superscript-III Reverse Transcriptase (Invitrogen, Carlsbad, CA). qPCR was performed using the Mx3000P (Stratagene, Santa Clara, CA), and the SYBR Premix Ex Taq (Takara, Kyoto, Japan). Cycle threshold (Ct) values were obtained using Stratagene Mx3000P software, and the Δ-Δ Ct method was used to calculate the relative fold change of RNA levels compared with a calibrator sample (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). Primer sequences are listed in supplemental Table 1. Transcript levels of apoptosis-related genes were assayed using the Mouse Apoptosis 96 StellARray qPCR array (Bar Harbor BioTechnology, Trenton, ME). The TissueScan Lymphoma Tissue qPCR Panel I (LYRT101; Origene, Rockville, MD) complementary DNA (cDNA) array was run according to the manufacturer’s instructions with primers for hEBI2 (supplemental Table 1) using a SYBR-green protocol. The cDNA samples on the plate had already been normalized against β-actin. Δ-Δ Ct values were calculated using the sample with the lowest Ct as the reference.
MACS sorting of cells
Single-cell suspensions of spleen tissue were enriched or depleted for CD19 or B220 by labeling with anti-CD19 or anti-B220 magnetic beads followed by MACS (Miltenyi Biotec, Bergisch Gladbach, Germany).
Antibodies
Monoclonal antibodies against CD3ε (145-2C11), CD4 (GK1.5), CD5 (53-7.3), CD8a (53-6.7), CD19 (6D5), CD21/CD35 (7E9), CD23 (B3B4), CD38 (90), B220 (RA3-6B2), CD69 (H1.2F3), CD80 (16-10A1), FAS (15A7), GL7 (GL7), IgM (R MM-1), IgD (11-26c.2a), and Ki-67 (SolA15) were obtained from BioLegend (San Diego, CA) whereas CD93 (AA4.1) and CD138 (281-2) were obtained from BD Biosciences (San Jose, CA). The antibodies were directly conjugated to Pacific Blue, fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE/Cy7, PE-CF594, allophycocyanin, or allophycocyanin/Cy7.
Flow cytometry (FACS)
Single-cell suspensions from spleen, thymus, kidney, lungs, and liver tissue as well as peritoneal lavage and EDTA-treated blood were red blood cell lysed using Gey solution. Cell suspensions were prepared in cold phosphate-buffered saline (PBS; without Ca2+ and Mg2+) supplemented with 1% bovine serum albumin, 10% rat serum, and 0.1% sodium azide. Cells were then stained with appropriate concentrations of the antibodies (Abs) named in the previous section in a volume of 100 μL and incubated at 4°C in the dark for 20 minutes. After washing twice with PBS supplemented with 1% sodium azide, cells were fixed in 1% paraformaldehyde and analyzed using the MoFlo Astrios (Beckman Coulter, Brea, CA). Events (105) were collected and analyzed by FlowJo VX software. For cell-sorting experiments, 5 × 106 cells were stained and sorted using the MoFlo Astrios (Beckman Coulter).
Immunization
Mice were immunized subcutaneously with 100 μg of 2,4-dinitrophenyl-ovalbumin (DNP-OVA) (Alpha Diagnostic International, San Antonio, TX) in alum. Three and 6 weeks after the first immunization the mice received booster immunizations with 100 μg of DNP-OVA in alum. Two weeks after each immunization a blood sample was taken and used for serum isolation. Two weeks after the second boost the mice were euthanized and used for serum isolation and B-cell subset analyses by FACS.
ELISA
Anti-DNP IgM and IgG levels in serum from immunized IgH-hEBI2 and WT mice were titrated by sandwich enzyme-linked immunosorbent assay (ELISA) using DNP-OVA–coated 96-well plates and rat anti-mouse IgM–horseradish peroxidase (HRP; Southern Biotech, Birmingham, AL) or rabbit anti-mouse IgG-HRP (Sigma-Aldrich, St. Louis, MO). The reactions were developed with 3,3′,5,5′-tetramethylbenzidine and H2O2, and the plates were read at 450 nm on an automated ELISA reader. Total serum IgG, IgM, and anti-double stranded DNA (dsDNA) IgM was assessed using the Mouse-IgG ELISA kit (Roche, Basel, Switzerland), the Mouse IgM ELISA kit (Sigma-Aldrich), and the Mouse Anti-dsDNA IgM ELISA kit (Alpha Diagnostic International). Serum samples were diluted 1/100 000, 1/1000, and 1/50, respectively, and the plates were read at 405 nm (IgG ELISA) or 450 nm (IgM and anti-dsDNA IgM ELISAs).
BrdU proliferation
Mice were treated with 0.8 mg/mL 5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) in their drinking water for 24 hours or 8 days. BrdU solution was prepared in sterile water, protected from light exposure, and changed daily. Single-cell suspensions were stained as described in the previous section with antibodies against B220 and CD19 followed by permeabilization and staining with anti-BrdU-PE (BD Biosciences). The fraction of BrdU-labeled B cells were assayed by use of FACS.
Histology
Tissues were fixed in 10% neutral-buffered formalin (Sigma-Aldrich) and after 2 days were transferred to 70% ethanol. Paraffin-embedded tissues were sectioned and stained with hematoxylin and eosin (H&E).
PCR assay for clonality and somatic hypermutations
For the preparation of high-molecular-weight DNA, spleen tissues were processed with the QIAamp DNA mini kit (Qiagen). The Expand High Fidelity PCR System (Roche) was used on 100-ng aliquots of genomic DNA to detect clonal immunoglobulin rearrangements. Hot-start 1-round amplification was performed with primers listed in supplemental Table 2 and the following program: initial denaturation at 96°C for 5 minutes with the reaction subsequently being held at 80°C for the addition of polymerase mix (1.75 units per 50-mL reaction). This was followed by 30 cycles of template denaturation at 95°C for 15 seconds, primer annealing at 62°C for 15 seconds, and primer extension at 68°C for 4 minutes in the first cycle with a progressive prolongation of the extension time by 20 seconds for each of the subsequent cycles.
Fragments were gel purified and cloned using the TOPO TA kit (Invitrogen). Plasmid DNA was sequenced using BigDye terminator. Sequence alignment was performed using IgBLAST (http://www.ncbi.nlm.nih.gov/igblast).
Adoptive transfer
Spleen cells from 16- to 18-month-old B6-WT and B6-IgH-hEBI2 mice were resuspended in PBS, and injected IV into the lateral tail vein (50 × 106 cells in 300-μL total volume) of B6-ν/ν mice. After 4 weeks, blood samples were taken to determine the presence of the B220low/− phenotype by FACS. Mice were euthanized after 8 weeks and examined for splenomegaly, the presence of the B220low/− phenotype cells, and histology of the spleen.
IL-10 blockade
IgH-hEBI2 and age-matched WT mice were injected intraperitoneally with 0.2 mg of rat anti-interleukin-10 (IL-10) (JES5-2A5; BioXCell, West Lebanon, NH) or isotype control rat IgG (HRPN; BioXCell) antibodies every second day for 3 weeks. Blood samples of all groups were taken at days 0, 8, 14, and 22 of initial injection and analyzed by FACS to assess cell counts and proliferation by Ki-67 staining.
Western blot
B220-expressing spleen cells were isolated using MACS sorting as described in the previous section followed by nuclear protein extraction and/or lysis using the Nuclear Extraction kit (Millipore, Billerica, MA). Equal amounts of protein were loaded on each lane, separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and blotted with the specific antibodies. The antibodies against c-Myc (C19) and BCL-2 (N19) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). β-actin (Cell Signaling) was used as a loading control. The bands were detected by use of goat anti-rabbit IgG-HRP (Sigma-Aldrich) and developed with SuperSignal West Pico Chemiluminescent Reagent (Thermo Scientific, Waltham, MA).
Apoptosis assays
Freshly isolated spleen cells were cultured in 1 mL of RPMI 1640 supplemented with 10% fetal calf serum at 5 × 106 cells/mL in 6-well plates. Cells were incubated with varying concentrations of etoposide for 24 hours and washed and stained with anti-B220. The cells were then washed and stained with propidium iodide (5 μg/mL; Sigma-Aldrich). Numbers of apoptotic cells were determined by FACS.
T-cell depletion
IgH-hEBI2 and WT mice were injected intraperitoneally every 3 to 4 days for 4 weeks with a cocktail of 200 mg of the following depleting antibodies: anti-CD4, anti-CD8, and anti-Thy-1. Blood samples were taken once a week and used for FACS to determine depletion of CD4+ and CD8+ cells. After 4 weeks, the mice were killed and the spleen sizes determined and single-cell suspensions from spleen tissue were used for FACS to determine depletion of CD4+ and CD8+ cells.
FISH
Fluorescence in situ hybridization (FISH) was performed on formalin-fixed paraffin-embedded tissue sections according to standard protocols (http://www.ihcworld.com/_protocols/lab_protocols/ried-lab-protocols.htm) using gene-specific probes for IgH and c-Myc. Probes were labeled with biotin or digoxigenin by nick translation (Roche). Detection was done with anti-digoxigenin FITC-labeled Fab fragments (Roche) and streptavidin–Alexa Fluor 568 (Invitrogen). Slides were counterstained and coverslipped using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Images were acquired using an Olympus IX81 fluorescent microscope and processed using Slidebook software (v.5.0.25; Intelligent Imaging Innovations).
Statistical analysis
Data were analyzed using the 2-tailed Student t test, 1-way analysis of variance, or the nonparametric Mann-Whitney test and the survival curves were compared using the log-rank test. A P value < .05 was considered statistically significant (*P < .05; **P < .01 ; ***P < .001; and ****P < .0001).
Results
Reduced GC responses in mice with hEBI2-expressing B cells
qPCR analysis of IgH-hEBI2 mice showed that hEBI2 was expressed primarily in CD19+ B cells and, consequently, was most abundant in the spleen (supplemental Figure 1A-B). Some expression was observed in the thymus (supplemental Figure 1B) and in the splenic CD19-depleted cell population (supplemental Figure 1A), which could be due to low expression in thymocytes as reported for other genes controlled by the IgH intronic enhancer.20,21
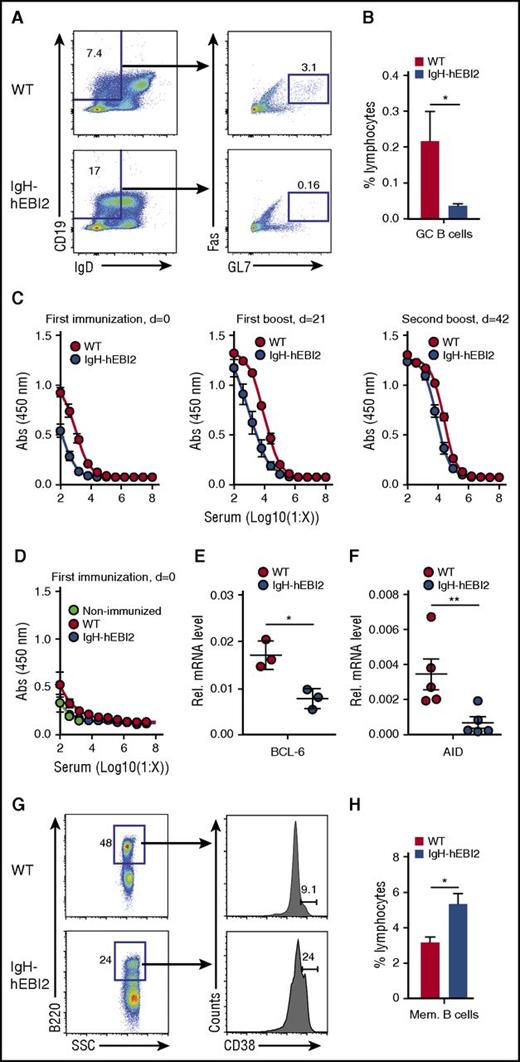
To investigate the GC development in IgH-hEBI2 mice, we first used flow cytometry to quantify GC B cells in the spleen of young adult (13-week-old) IgH-hEBI2 mice and found reduced levels of GC cells as compared with WT mice (Figure 1A-B). To investigate the ability of hEBI2-expressing B cells to participate in GC reactions, we immunized WT and IgH-hEBI2 mice with the T-cell–dependent antigen DNP-OVA. IgH-hEBI2 mice had reduced anti-DNP responses at 2 weeks after the first immunization and 2 weeks after the first boost (Figure 1C). Levels of anti-DNP IgM after the first immunization were not significantly reduced (Figure 1D). These data suggest that hEBI2-expressing B cells, similarly to what has previously been shown for B cells with enforced expression of murine EBI2,5 have a reduced ability to participate in the GC-dependent antibody response. Accordingly, transcript levels for both BCL-6, which is essential to GC reactions,22 and the enzyme AID, which is required for somatic hypermutation and class switch recombination,23 were reduced in IgH-hEBI2 mice (Figure 1E-F). After a second boost, the levels of anti-DNP IgG were similar in IgH-hEBI2 and WT mice (Figure 1C), likely due to gradual acquisition of high-affinity antibodies by memory B cells. Indeed, the size of the memory B-cell compartment in IgH-hEBI2 mice was increased compared with that in WT mice after a second DNP-OVA boost (Figure 1G-H).
Reduced GC response in IgH-hEBI2 mice. (A). Gating scheme used to define the splenic CD19+IgDlowFas+GL7+ GC B-cell population. (B) Subsets of splenic GC B cells of young (13 weeks) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of GC B cells are given as percentage of total lymphocytes. Data are mean ± standard error of the mean (SEM) of data from 5 to 6 mice. *P < .05 by the Student t test. (C). IgH-hEBI2 and WT mice were immunized with DNP-OVA precipitated in alum followed by boosters 21 and 42 days (d) later. The concentration of serum anti-DNP-OVA IgG1 in the mice was determined by ELISA in serum samples taken 2 weeks after the indicated time points. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT mice. P values for log 50% inhibitory concentration (IC50) values are .0037 and <.0001 for serum concentrations after the first immunization and the first boost, respectively. (D). IgH-hEBI2 and WT mice were immunized with DNP-OVA precipitated in alum. The concentration of serum anti-DNP-OVA IgM was determined by ELISA in serum samples taken 2 weeks after immunization. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT immunized mice and 3 nonimmunized control mice. (E). qPCR analysis of BCL-6 expression in spleen cells from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 3 mice. *P < .05 by nonparametric Mann-Whitney test. (F). qPCR analysis of AID expression in spleen cells from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 5 mice. **P < .01 by nonparametric Mann-Whitney test. (G). Gating scheme used to define splenic B220+CD38+ memory B cells in DNP-OVA immunized mice. (H). Number of memory B cells in spleens of DNP-OVA immunized WT and IgH-hEBI2. Mice were euthanized 2 weeks after the second booster immunization and spleens were used for FACS. The number of memory cells is given as percentage of total lymphocytes. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT mice. *P < .05 by Student t test. AID, activation-induced cytidine deaminase; SSC, side scatter.
Reduced GC response in IgH-hEBI2 mice. (A). Gating scheme used to define the splenic CD19+IgDlowFas+GL7+ GC B-cell population. (B) Subsets of splenic GC B cells of young (13 weeks) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of GC B cells are given as percentage of total lymphocytes. Data are mean ± standard error of the mean (SEM) of data from 5 to 6 mice. *P < .05 by the Student t test. (C). IgH-hEBI2 and WT mice were immunized with DNP-OVA precipitated in alum followed by boosters 21 and 42 days (d) later. The concentration of serum anti-DNP-OVA IgG1 in the mice was determined by ELISA in serum samples taken 2 weeks after the indicated time points. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT mice. P values for log 50% inhibitory concentration (IC50) values are .0037 and <.0001 for serum concentrations after the first immunization and the first boost, respectively. (D). IgH-hEBI2 and WT mice were immunized with DNP-OVA precipitated in alum. The concentration of serum anti-DNP-OVA IgM was determined by ELISA in serum samples taken 2 weeks after immunization. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT immunized mice and 3 nonimmunized control mice. (E). qPCR analysis of BCL-6 expression in spleen cells from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 3 mice. *P < .05 by nonparametric Mann-Whitney test. (F). qPCR analysis of AID expression in spleen cells from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 5 mice. **P < .01 by nonparametric Mann-Whitney test. (G). Gating scheme used to define splenic B220+CD38+ memory B cells in DNP-OVA immunized mice. (H). Number of memory B cells in spleens of DNP-OVA immunized WT and IgH-hEBI2. Mice were euthanized 2 weeks after the second booster immunization and spleens were used for FACS. The number of memory cells is given as percentage of total lymphocytes. The results are mean ± SEM of data from 10 IgH-hEBI2 and 7 WT mice. *P < .05 by Student t test. AID, activation-induced cytidine deaminase; SSC, side scatter.
Increased number of B1 cells in IgH-hEBI2 mice
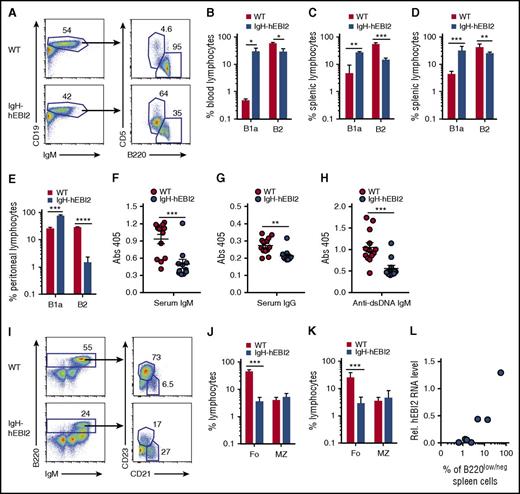
To evaluate the effect of hEBI2 expression on the distribution of B-cell subsets, we examined blood cells from IgH-hEBI2 and WT mice by flow cytometry (FACS). These data revealed that hEBI2 expression shifts the B-cell subset toward a B1a CD19+/B220low/−/CD5+ subset (Figure 2A-B). This finding was mirrored in analyses of spleen cells, which revealed that both young adult (12-15 weeks) (Figure 2C) and old (16-18 months) (Figure 2D) IgH-hEBI2 mice had expanded populations of B1a cells. B1a cells are normally almost absent in the spleen and instead locate to the peritoneum and pleural cavities.4 Flow analysis of B-cell subsets in the peritoneum revealed that around 80% of the IgH-hEBI2 B cells were B1a cells compared with only 25% in WT mice (Figure 2E). To further investigate the consequences of increased numbers of B1a B cells in IgH-hEBI2 mice, we measured the serum levels of IgM, IgG, and IgM antibodies against dsDNA as B1 B cells are known to secrete high levels of polyreactive immunoglobulins (such as antibodies against dsDNA). However, as shown in Figure 2F-H, we found lower levels of IgM, IgG, and anti-dsDNA IgM, indicating that, although rich in B1a cells, the IgH-hEBI2 mice are characterized by pronounced hypoimmunoglobulinemia.
Increased number of B1a cells in IgH-hEBI2 mice. (A). Gating scheme used to define CD19+IgM+B220lowCD5+ B1a and CD19+IgM+B220+CD5− B2 cell populations in spleen and blood. (B) B1a and B2 B-cell subsets in blood samples from 6- to 9-month-old WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The number of cells in the B-cell subsets is given as percentage of total leukocytes. The results are mean ± SEM of data from 3 mice. *P < .05 by Student t test. (C-D) Subsets of splenic B1a and B2 B cells of young (12-15 weeks) (C) and old (16-18 months) (D) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of cells in the B1a and B2 B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 3 to 9 mice. **P < .01 and ***P < .001 by Student t test. (E). Subsets of B1 and B2 B cells in the peritoneum of young (13 weeks) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of cells in the B1a and B2 B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 5 mice. ***P < .001 and ****P < .0001 by Student t test. (F-H) The concentration of serum IgM (F), IgG (G), and anti-dsDNA IgM (H) in IgH-hEBI2 and WT mice was determined by ELISA. The results are mean ± SEM of data from 13 IgH-hEBI2 and 12 WT mice. **P < .01 and ***P < .001 by Student t test. (I) Gating scheme used to define splenic B220+IgM+CD21+CD23− MZ and B220+IgM+CD21−CD23+ follicular B-cell subsets. (J-K) Subsets of splenic follicular (Fo.) and MZ B cells of young (12-15 weeks) (J) and old (16-18 months) (K) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel I. Cells were gated on lymphocytes. The numbers of cells in the follicular and MZ B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 3 to 9 mice. ***P < .001 by Student t test. (L) Correlation of the numbers of CD19+/B220low/− B cells as determined by FACS and hEBI2 expression as determined by qPCR in 7 IgH-hEBI2 founder mice. Pearson correlation coefficient is 0.96 with P = .0005.
Increased number of B1a cells in IgH-hEBI2 mice. (A). Gating scheme used to define CD19+IgM+B220lowCD5+ B1a and CD19+IgM+B220+CD5− B2 cell populations in spleen and blood. (B) B1a and B2 B-cell subsets in blood samples from 6- to 9-month-old WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The number of cells in the B-cell subsets is given as percentage of total leukocytes. The results are mean ± SEM of data from 3 mice. *P < .05 by Student t test. (C-D) Subsets of splenic B1a and B2 B cells of young (12-15 weeks) (C) and old (16-18 months) (D) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of cells in the B1a and B2 B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 3 to 9 mice. **P < .01 and ***P < .001 by Student t test. (E). Subsets of B1 and B2 B cells in the peritoneum of young (13 weeks) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel A. Cells were gated on lymphocytes. The numbers of cells in the B1a and B2 B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 5 mice. ***P < .001 and ****P < .0001 by Student t test. (F-H) The concentration of serum IgM (F), IgG (G), and anti-dsDNA IgM (H) in IgH-hEBI2 and WT mice was determined by ELISA. The results are mean ± SEM of data from 13 IgH-hEBI2 and 12 WT mice. **P < .01 and ***P < .001 by Student t test. (I) Gating scheme used to define splenic B220+IgM+CD21+CD23− MZ and B220+IgM+CD21−CD23+ follicular B-cell subsets. (J-K) Subsets of splenic follicular (Fo.) and MZ B cells of young (12-15 weeks) (J) and old (16-18 months) (K) WT and IgH-hEBI2 mice were analyzed by FACS as shown in panel I. Cells were gated on lymphocytes. The numbers of cells in the follicular and MZ B-cell subsets are given as percentage of total lymphocytes. Data are mean ± SEM of data from 3 to 9 mice. ***P < .001 by Student t test. (L) Correlation of the numbers of CD19+/B220low/− B cells as determined by FACS and hEBI2 expression as determined by qPCR in 7 IgH-hEBI2 founder mice. Pearson correlation coefficient is 0.96 with P = .0005.
The dominance of B1a cells in the peritoneal compartment and the unusual occurrence of high numbers of these cells in the blood and spleen would imply a reduction of cells from other normal B-cell compartments. In agreement with this hypothesis, the B2 CD19+/B220hi compartment was reduced with fewer follicular B cells (Figure 2I-K). In contrast, IgH-hEBI2 mice did not differ from WT mice in the frequencies of MZ B cells, which share many functional properties with B1 cells24 (Figure 2I-K). The number of transitional B cells, activated B cells, and plasmablasts were similar or reduced in IgH-hEBI2 mice (supplemental Figure 2A-C). hEBI2 transcripts were present in all B-cell subsets of IgH-hEBI2 mice (supplemental Figure 3). Further FACS analyses showed that the frequencies of dendritic cells, granulocytes, T cells, and natural killer cells were the same in young adult (12-15 weeks), 12-, and 16-month-old IgH-hEBI2 and WT mice (supplemental Figure 4A-F). The study of 6 new founder mice confirmed that the shift toward CD19+/B220low/− B1 cells in IgH-hEBI2 mice correlated positively with expression levels of hEBI2 (Figure 2L).
Proliferation and lymphoma development in IgH-hEBI2 mice
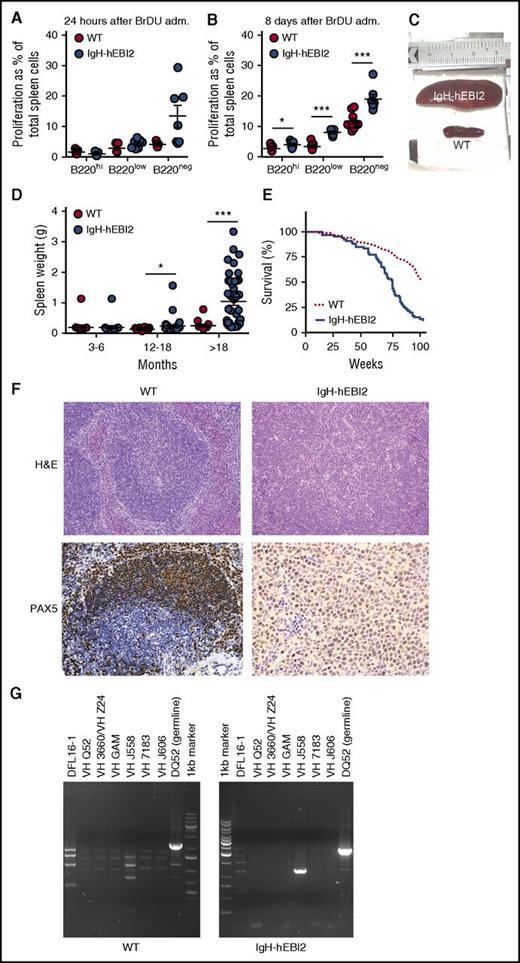
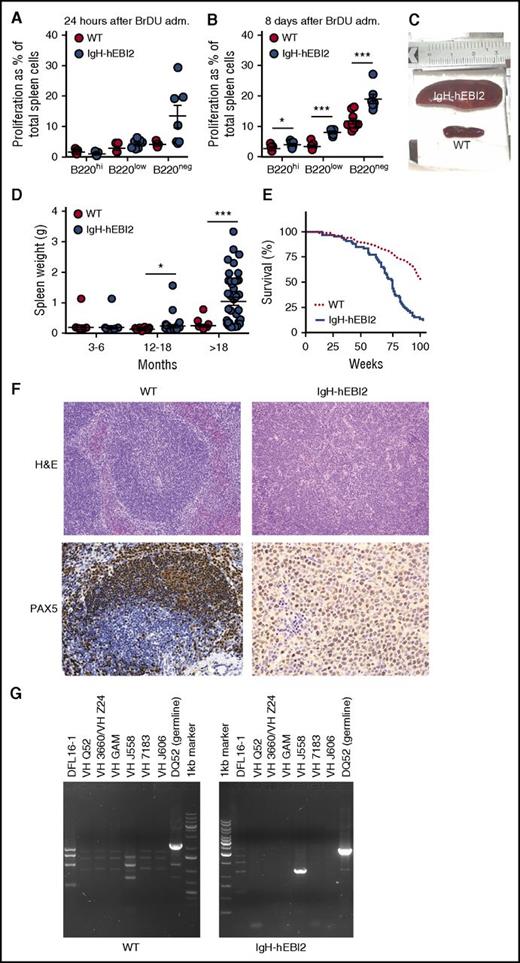
To assess whether the increased number of B1a CD19+/B220low/−/CD5+ B cells in IgH-hEBI2 mice was due to increased proliferation, we tested spleen cells for in vivo incorporation of BrdU in young adult (12-15 weeks) mice. After 24 hours, there was a tendency toward an increased frequency of BrdU+ CD19+/B220low and CD19+/B220− B1a B cells in IgH-hEBI2 mice (P values of .12 and .06, respectively) (Figure 3A). At 8 days, the frequencies of BrdU+ B1a B cells as well as BrdU+ CD19+/B220hi B2 B cells were significantly increased in IgH-hEBI2 mouse spleens (Figure 3B), suggesting that the increased proliferation can be directly caused by hEBI2 expression and not only by changes in B-cell subset distribution. These data are consistent with previous ex vivo findings where stimulation with α-IgM resulted in increased proliferation of splenic cells from IgH-hEBI2 mice.12
Increased proliferation of B cells and lymphoma development in IgH-hEBI2 mice. (A-B). BrdU incorporation after 24 hours (A) and accumulation after 8 days (B) in CD19+/B220hi, CD19+/B220low, and CD19+/B220− IgH-hEBI2 and WT spleen cells as detected by FACS. The proliferation is shown as percentage of BrdU incorporation in the total spleen cell population. The results are mean ± SEM of data from 5 to 9 young (12-15 weeks) mice. *P < .05 and ***P < .001 by Student t test. (C). Picture of spleens from age-matched IgH-hEBI2 (top spleen) and WT (bottom spleen) animals. The spleens shown are representative of cases from 16-month-old animals. (D) Spleen weight in IgH-hEBI2 and WT mice over time. The results are data from 14 to 40 mice. *P < .05 and ***P < .001 by Student t test. (E) Kaplan-Meier survival plot of IgH-hEBI2 and WT mice. P < .001. (F) Representative histologic sections from an IgH-hEBI2 lymphoma mouse and an age-matched WT mouse (16 months old). All samples are stained with H&E (original magnification ×100) or PAX5 (B-cell marker) (original magnification ×200 and ×400 for WT and IgH-hEBI2 sections, respectively). (G) PCR analysis of clonality in IgH-hEBI2 mice. The analysis was done with JH4-1 reverse primer for all lanes and forward primers specific for V or D segments as noted above the individual lanes.
Increased proliferation of B cells and lymphoma development in IgH-hEBI2 mice. (A-B). BrdU incorporation after 24 hours (A) and accumulation after 8 days (B) in CD19+/B220hi, CD19+/B220low, and CD19+/B220− IgH-hEBI2 and WT spleen cells as detected by FACS. The proliferation is shown as percentage of BrdU incorporation in the total spleen cell population. The results are mean ± SEM of data from 5 to 9 young (12-15 weeks) mice. *P < .05 and ***P < .001 by Student t test. (C). Picture of spleens from age-matched IgH-hEBI2 (top spleen) and WT (bottom spleen) animals. The spleens shown are representative of cases from 16-month-old animals. (D) Spleen weight in IgH-hEBI2 and WT mice over time. The results are data from 14 to 40 mice. *P < .05 and ***P < .001 by Student t test. (E) Kaplan-Meier survival plot of IgH-hEBI2 and WT mice. P < .001. (F) Representative histologic sections from an IgH-hEBI2 lymphoma mouse and an age-matched WT mouse (16 months old). All samples are stained with H&E (original magnification ×100) or PAX5 (B-cell marker) (original magnification ×200 and ×400 for WT and IgH-hEBI2 sections, respectively). (G) PCR analysis of clonality in IgH-hEBI2 mice. The analysis was done with JH4-1 reverse primer for all lanes and forward primers specific for V or D segments as noted above the individual lanes.
The preference for CD5+ B1a cell expansion by hEBI2 overexpression combined with increased proliferation most likely contributed to a CLL-like splenomegaly, which was observed to develop in ∼30% of IgH-hEBI2 mice older than 12 months and for 90% of mice older than 18 months (Figure 3C-D; Table 1). As a consequence, the survival rate of IgH-hEBI2 mice was significantly reduced (P < .001) (Figure 3E). Histologic studies of enlarged spleens revealed that the mice were dying with lymphomas with a consistent infiltration of small and large lymphocytes and disrupture of the normal structure (Figure 3F), but the lymphomas did not appear until the age of 12 months (Figure 3D), long after highly proliferative B cells were already present (Figure 3A-B). The percentages of total B220+ cells and B220low/− B1 cells in spleens of IgH-hEBI2 mice increased from 3 to 6 months to 12 months of age and decreased in mice older than 18 months (supplemental Figure 5A-B). Staining for PAX5 confirmed that the lymphomas were of B-cell origin (Figure 3F). To examine lymphoma clonality, we performed PCR analyses of IgH rearrangements and identified monoclonal expansions of B cells (Figure 3G). Analyses of VDJ sequences revealed no somatic mutations, suggesting that the lymphomas originated from B cells that had not undergone GC passage (supplemental Table 3). This is consistent with the finding that most B cells in aged IgH-hEBI2 mice belong to the B1a subset (Figure 2A,D).
Adoptive transfer of lymphomas from IgH-hEBI2 mice
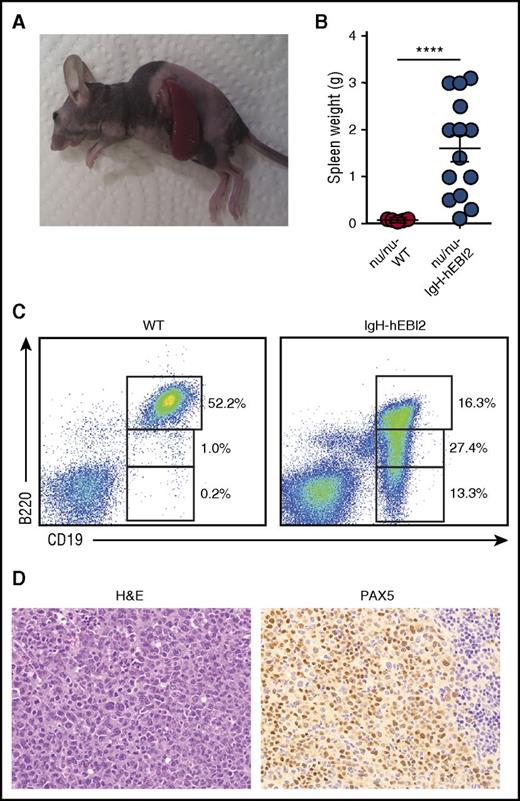
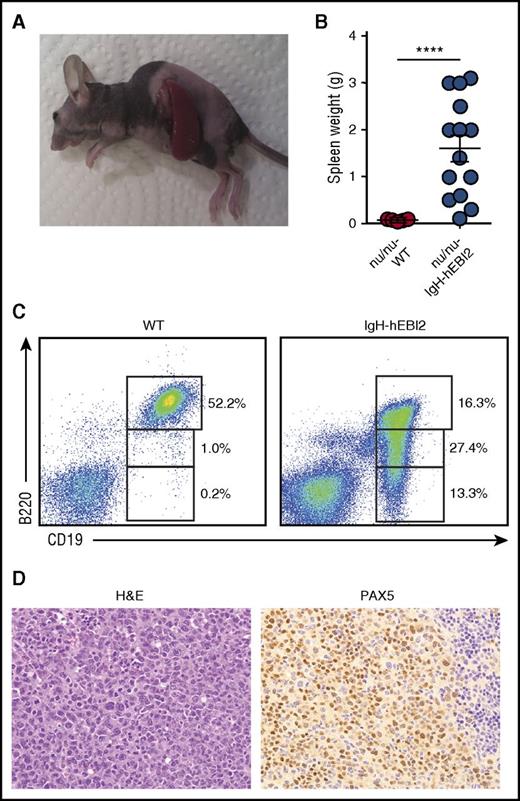
A hallmark of cancers is the ability to grow after transplantation into immunodeficient hosts. Spleen cells from IgH-hEBI2 mice with lymphomas and WT mice were transplanted to immunodeficient nude (ν/ν) mice. After 8 weeks, all of the ν/ν mice, which had received cells from IgH-hEBI2 mice, had developed lymphomas whereas recipients of cells from WT mice did not (Figure 4A-B). The transplanted tumors had the same CD19+/B220low/− B1 cell phenotype as B cells from the donor IgH-hEBI2 mice (Figure 4C) and had the same histopathologic features as the original lymphomas (Figure 4D). WT mice injected with lymphomas from IgH-hEBI2 mice did not develop splenomegaly (data not shown). The inability of IgH-hEBI2 lymphoma cells to be transplanted into WT mice could be due to a number of reasons, including immune surveillance as previously reported for LMP1-transgenic mice.25
Lymphoma cells from IgH-hEBI2 animals can be transferred to and survive in nude mice. (A) Picture of a B6-ν/ν mouse injected with lymphoma cells from a 16-month-old IgH-hEBI2 mouse. The picture is representative of mice injected with lymphoma spleen cells from IgH-hEBI2 mice. (B) Spleen weights of B6-ν/ν mice injected with spleen cells from age-matched control WT mice and lymphoma IgH-hEBI2 mice. The results are data from 10 to 14 mice. ****P < .0001 by Student t test. (C) Representative plot of surface expression of B220 and CD19 on spleen cells from B6 nude mice injected with lymphoma cells from IgH-hEBI2 mice and spleen cells from WT mice and quantification of CD19+/B220hi, CD19+/B220low, and CD19+/B220− B-cell subsets as determined by FACS. (D) Representative histologic section from an B6-ν/ν mouse injected with spleen cells from a IgH-hEBI2 lymphomatic mice. The sample is stained with H&E or PAX5 (B-cell marker) (original magnification ×400).
Lymphoma cells from IgH-hEBI2 animals can be transferred to and survive in nude mice. (A) Picture of a B6-ν/ν mouse injected with lymphoma cells from a 16-month-old IgH-hEBI2 mouse. The picture is representative of mice injected with lymphoma spleen cells from IgH-hEBI2 mice. (B) Spleen weights of B6-ν/ν mice injected with spleen cells from age-matched control WT mice and lymphoma IgH-hEBI2 mice. The results are data from 10 to 14 mice. ****P < .0001 by Student t test. (C) Representative plot of surface expression of B220 and CD19 on spleen cells from B6 nude mice injected with lymphoma cells from IgH-hEBI2 mice and spleen cells from WT mice and quantification of CD19+/B220hi, CD19+/B220low, and CD19+/B220− B-cell subsets as determined by FACS. (D) Representative histologic section from an B6-ν/ν mouse injected with spleen cells from a IgH-hEBI2 lymphomatic mice. The sample is stained with H&E or PAX5 (B-cell marker) (original magnification ×400).
Immune surveillance and T-cell activation in IgH-hEBI2 mice
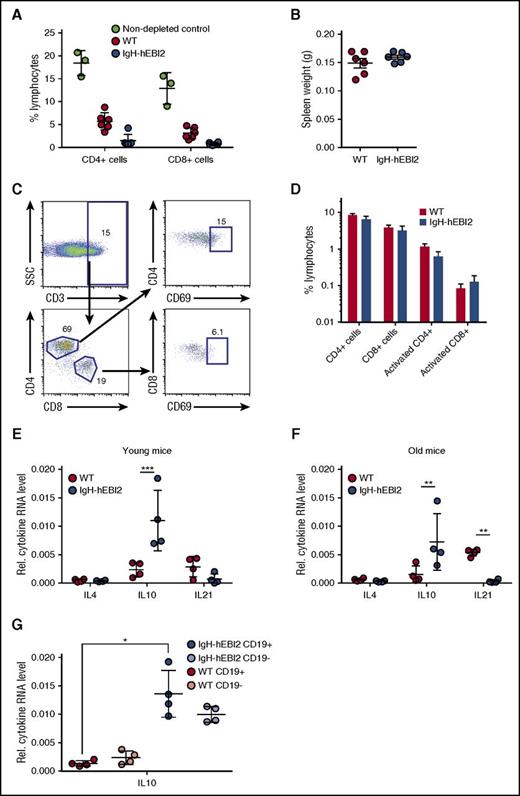
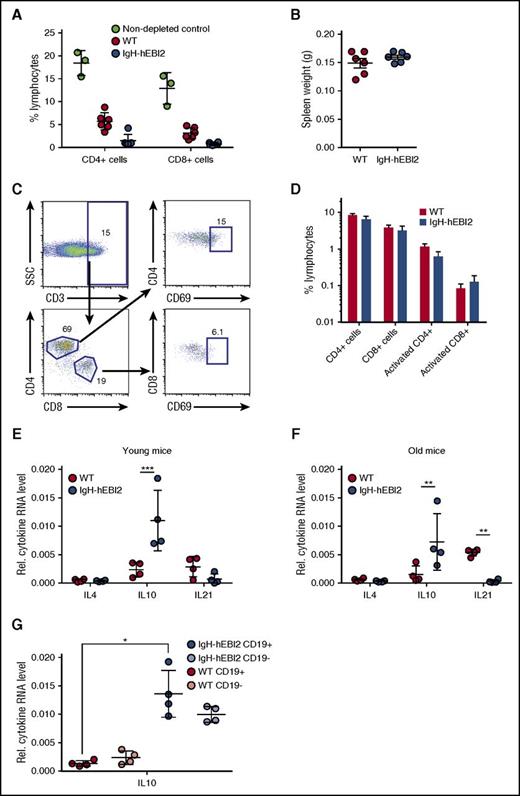
To test whether immune surveillance played a role in delaying disease progression in IgH-hEBI2 mice, we injected WT and IgH-hEBI2 mice with depleting antibodies against CD4, CD8, and Thy1. Although FACS analyses confirmed depletion of CD4+ and CD8+ cells (Figure 5A), the antibody-injected IgH-hEBI2 (and WT) mice did not develop splenomegaly within the timeframe of the experiment (4 weeks) (Figure 5B). Flow analyses on aged mice revealed that neither the CD4+ or CD8+ T-cell subsets were expanded in IgH-hEBI2 mice (supplemental Figure 4A-B) and further analyses showed that neither of these cell subsets were activated in aged IgH-hEBI2 mice (Figure 5C-D).
Splenic T cells are not activated in IgH-hEBI2 mice, but increased levels of the B1 B-cell–secreted IL-10 are observed. (A) CD4+ and CD8+ T cells were depleted by injection of antibodies against CD4, CD8, and Thy1. The percentage of CD4+ and CD8+ cells in WT and IgH-hEBI2 mice were measured by FACS. The results are given as percentage of total spleen cells and are mean ± SEM of data from 6 T-cell–depleted IgH-hEBI2 and WT mice and 3 nondepleted control mice. (B) Spleen weight of WT and IgH-hEBI2 mice after 4 weeks of T-cell–depleting injections with antibodies against CD4, CD8, and Thy1. The results are given as mean ± SEM of data from 6 T-cell–depleted IgH-hEBI2 and WT mice. (C) Gating scheme for activated T cells. (D) Spleen cells from 12- to 15-week-old WT and IgH-hEBI2 mice were analyzed for the presence of the T-cell activation marker CD69 by FACS. Numbers indicate the percentage of cells falling into each gate as shown in panel C. The results are mean ± SEM of data from 5 WT and 6 IgH-hEBI2 mice. (E-F) qPCR analysis of IL-4, IL-10, and IL-21 expression in spleen cells from young (12-15 weeks) (E) and old (16-18 months) (F) IgH-hEBI2 and WT mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 4 mice. **P < .01 and ***P < .001 by the nonparametric Mann-Whitney test. (G) qPCR analysis of IL-10 expression in CD19-enriched and -depleted spleen samples from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 4 mice. *P < .05 by the nonparametric Mann-Whitney test.
Splenic T cells are not activated in IgH-hEBI2 mice, but increased levels of the B1 B-cell–secreted IL-10 are observed. (A) CD4+ and CD8+ T cells were depleted by injection of antibodies against CD4, CD8, and Thy1. The percentage of CD4+ and CD8+ cells in WT and IgH-hEBI2 mice were measured by FACS. The results are given as percentage of total spleen cells and are mean ± SEM of data from 6 T-cell–depleted IgH-hEBI2 and WT mice and 3 nondepleted control mice. (B) Spleen weight of WT and IgH-hEBI2 mice after 4 weeks of T-cell–depleting injections with antibodies against CD4, CD8, and Thy1. The results are given as mean ± SEM of data from 6 T-cell–depleted IgH-hEBI2 and WT mice. (C) Gating scheme for activated T cells. (D) Spleen cells from 12- to 15-week-old WT and IgH-hEBI2 mice were analyzed for the presence of the T-cell activation marker CD69 by FACS. Numbers indicate the percentage of cells falling into each gate as shown in panel C. The results are mean ± SEM of data from 5 WT and 6 IgH-hEBI2 mice. (E-F) qPCR analysis of IL-4, IL-10, and IL-21 expression in spleen cells from young (12-15 weeks) (E) and old (16-18 months) (F) IgH-hEBI2 and WT mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 4 mice. **P < .01 and ***P < .001 by the nonparametric Mann-Whitney test. (G) qPCR analysis of IL-10 expression in CD19-enriched and -depleted spleen samples from 12- to 15-week-old IgH-hEBI2 and WT mice relative to the expression of the control gene, GAPDH. The results are mean ± SEM of data from 4 mice. *P < .05 by the nonparametric Mann-Whitney test.
To assay whether the increased proliferation of IgH-hEBI2 B cells was induced by B-cell stimulatory cytokines that could be secreted by T cells, we measured the transcript levels of IL-4 (expressed by T helper cells), IL-10 (expressed by activated T cells and regulatory T cells), and IL-21 (expressed by CD4 T cells). Neither IL-4 or IL-21 were increased in either young adult (12-15 weeks) nor old (16-18 months) IgH-hEBI2 mice (Figure 5E-F). Indeed, in old IgH-hEBI2 mice, IL-21 transcripts were significantly reduced. In contrast, transcripts of IL-10 were increased in both young adult and old mice (Figure 5E-F). As IL-10 can be produced by both T and B cells (but plays opposite roles in the 2 cell populations as it induces proliferation in B cells while inhibiting it in T cells26 ), we sorted splenocytes into CD19-enriched and -depleted fractions. Increased IL-10 transcripts were detected in both fractions (Figure 5G), not only consistent with the B1a B cells of IgH-hEBI2 mice as a major source of IL-10,27 but also suggesting that non-B cells produce increased amounts of IL-10 in IgH-hEBI2 mice. To test whether the increased IL-10 transcription in IgH-hEBI2 mice impacts the T-cell activity or the increased proliferation, we treated age-matched WT and IgH-hEBI2 mice with anti-IL-10 antibodies, thereby neutralizing IL-10. We find that there is no effect of neutralizing IL-10 on neither B1 and B2 B-cell numbers or proliferation in these cell subsets when comparing with isotype control-treated IgH-hEBI2 mice (supplemental Figure 6A,B,D,E), perhaps reflecting the indolent stage of CLL development in mice at 12 to 15 weeks of age, as only progressive cases of human CLL respond directly to IL-10 with increased proliferation.28 Notably, although the number of total and proliferating (Ki-67+) CD3+ T cells in IgH-hEBI2 mice does not increase upon IL-10 neutralization when comparing to isotype control–treated IgH-hEBI2 mice (supplemental Figure 6C,F), IL-10 blockade in WT mice did increase the number of proliferating CD3+ T cells when comparing to isotype control–treated WT mice after 3 weeks of antibody treatment (supplemental Figure 6C,F). This finding demonstrates the effect of the treatment.
Expression of cellular oncogenes in hEBI2-expressing B cells
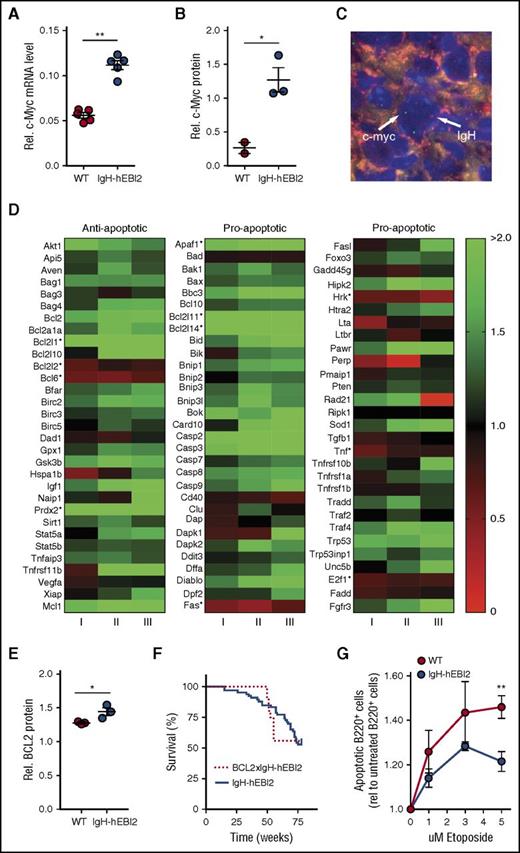
EBI2 has been shown to be upregulated upon EBV infection of B cells29 in addition to EBV+ posttransplant lymphoproliferative transplant samples.30 As cMyc is a major endogenous oncogene in many EBV-associated cancers,31 we performed qPCR and western blot analyses for cMyc RNA and protein expression in isolated B220+ cells from young adult (12-15 weeks) IgH-hEBI2 and WT mice. B220+ cells were used to directly compare the effect of hEBI2 expression and avoid effects attributed to differences in B-cell subset distribution. Expression of hEBI2 was associated with increased levels of cMyc transcripts and protein (Figure 6A-B; supplemental Figure 7A). FISH analyses on lymphoma samples revealed that this was not due to translocation of the cMyc gene to the IgH enhancer as seen in the EBV+ cancer Burkitt lymphoma31 (Figure 6C; individual figures are shown in supplemental Figure 7B).
Increased oncogene expression and resistance to apoptosis in IgH-hEBI2 mice. (A) qPCR analysis of c-Myc transcript levels in splenic B220+ cell populations from IgH-hEBI2 and WT mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 mice. **P < .01 by the nonparametric Mann-Whitney test. (B) Quantification of expression of c-Myc protein in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by western blot. The results are mean ± SEM of data from 2 to 3 mice. *P < .05 by Student t test. (C) FISH analysis of IgH and c-Myc localization in IgH-hEBI2 mice. The IgH allele is labeled with biotin and detected with streptavidin–AlexaFluor 568 (red). c-Myc is labeled with digoxygenin and detected with anti-Dig-FITC (green). The nuclei are stained with DAPI (blue). Image taken with a ×100 oil immersion objective magnification. (D) Heatmap of 93 anti- and proapoptotic genes and their expression in B220+ cells in young (12-15 weeks) IgH-hEBI2 in relation to the corresponding transcription in age-matched WT mice. Data shown from 3 IgH-hEBI2 mice. *P < .05 by Student t test. (E) Quantification of expression of BCL-2 protein in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by western blot. The results are mean ± SEM of data from 2 to 3 mice. *P < .05 by Student t test. (F) Kaplan-Meier plot of IgH-hEBI2 and BCL-2xIgH-hEBI2 mice. The curves show data from 66 (IgH-hEBI2) and 9 (BCL-2xIgH-hEBI2) mice. (G) Etoposide (topoisomerase II inhibitor) induced apoptosis in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by FACS with the late apoptotic marker propidium iodide. The results are given as fold increase in apoptotic cells compared with untreated cells and shown as mean ± SEM of data from 5 mice. **P < .01 by Student t test.
Increased oncogene expression and resistance to apoptosis in IgH-hEBI2 mice. (A) qPCR analysis of c-Myc transcript levels in splenic B220+ cell populations from IgH-hEBI2 and WT mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 mice. **P < .01 by the nonparametric Mann-Whitney test. (B) Quantification of expression of c-Myc protein in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by western blot. The results are mean ± SEM of data from 2 to 3 mice. *P < .05 by Student t test. (C) FISH analysis of IgH and c-Myc localization in IgH-hEBI2 mice. The IgH allele is labeled with biotin and detected with streptavidin–AlexaFluor 568 (red). c-Myc is labeled with digoxygenin and detected with anti-Dig-FITC (green). The nuclei are stained with DAPI (blue). Image taken with a ×100 oil immersion objective magnification. (D) Heatmap of 93 anti- and proapoptotic genes and their expression in B220+ cells in young (12-15 weeks) IgH-hEBI2 in relation to the corresponding transcription in age-matched WT mice. Data shown from 3 IgH-hEBI2 mice. *P < .05 by Student t test. (E) Quantification of expression of BCL-2 protein in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by western blot. The results are mean ± SEM of data from 2 to 3 mice. *P < .05 by Student t test. (F) Kaplan-Meier plot of IgH-hEBI2 and BCL-2xIgH-hEBI2 mice. The curves show data from 66 (IgH-hEBI2) and 9 (BCL-2xIgH-hEBI2) mice. (G) Etoposide (topoisomerase II inhibitor) induced apoptosis in spleen B220+ cells from IgH-hEBI2 and WT mice as determined by FACS with the late apoptotic marker propidium iodide. The results are given as fold increase in apoptotic cells compared with untreated cells and shown as mean ± SEM of data from 5 mice. **P < .01 by Student t test.
A previous study has proposed EBI2 as a negative regulator of innate immunity by inhibiting macrophage-induced apoptosis.32 We therefore assayed the expression of 93 anti- and proapoptotic genes in young adult IgH-hEBI2 and WT mice (Figure 6D). These data showed that there is no significant upregulation of antiapoptotic genes in IgH-hEBI2 compared with the upregulation of proapoptotic genes (supplemental Figure 7C). Nor is there a significant downregulation in antiapoptotic genes in IgH-hEBI2 compared with the downregulation of proapoptotic genes (supplemental Figure 7D). However, numerous genes, including MCL-1, that are implicated in lymphomagenesis were upregulated at the messenger RNA (mRNA) level. Also BCL-2, which is increased in many CLL patients,33 showed a tendency toward upregulation (P = .08) at the transcript level and a significant increase on protein level (Figure 6E; supplemental Figure 7E). Notably, crossing hEBI2-transgenic mice with BCL-2 transgenic mice did not reduce the survival rate more than what was observed with the hEBI2-transgenic mice (Figure 6F), hinting that a contributing mechanism behind the lymphoma development was already an antiapoptotic effect to which BCL-2 could not contribute further. To explore this, we tested whether hEBI2 inhibits apoptosis in B cells ex vivo. We treated purified B220+ splenic B cells from young adult (12-15 weeks) IgH-hEBI2 and WT mice with the topoisomerase II inhibitor, etoposide. Expression of hEBI2 was associated with reduced apoptosis at all concentrations tested (Figure 6G).
Relevance of EBI2 expression to CLL and other human lymphoid cancers
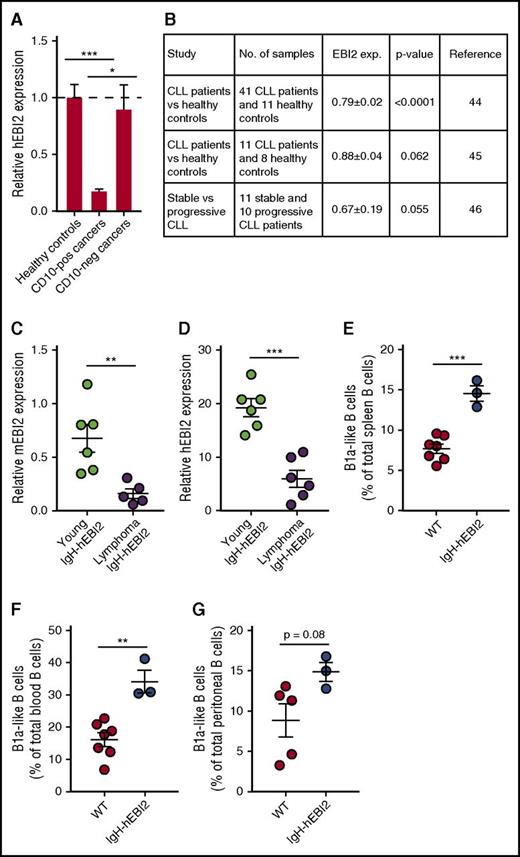
There is no direct link between EBI2 expression and human lymphoid cancers except for the link provided by this study and by the known ability of EBV infection to upregulate EBI2 and promote human cancer development.30 However, to address the potential importance of EBI2 for follicular and extrafollicular lymphoid cancers, we chose to assay EBI2 expression in a cohort of lymphomas with a lymphoma array from Origene and primers against hEBI2 and divided the data into CD10+ GC cancers (diffuse large B-cell lymphoma, follicular lymphoma, Hodgkin lymphoma) and CD10− non-GC cancers (MZ lymphoma and mantle cell lymphoma). We found no difference in EBI2 transcript levels between healthy controls and CD10− cancers, but we did see a large reduction in EBI2 transcript levels in CD10+ cancers compared with both healthy controls and CD10− cancers (Figure 7A), supporting the concept that EBI2 is downregulated in cancers that invade the GCs.5,6
Expression of EBI2 in lymphoma patients. (A) qPCR analysis of EBI2 transcript levels in CD10+ and CD10− cancers relative to the mean expression level of the healthy control samples. The cDNA samples in the array had been prenormalized to β-actin. The results are mean ± SEM of data from 6 to 14 samples. *P < .05 and ***P < .001 by Student t test. (B) Expression data on EBI2 expression in CLL patients compared with healthy controls. Expression data from Gene Expression Omnibus (GEO) profiles reprinted from Gutierrez et al,45 Gutiérrez et al46 with permission, and Fält et al47 with permission. (C) qPCR analysis of mEBI2 transcript levels in spleen cells from young (12-15 weeks) and older, lymphoma IgH-hEBI2 mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 to 6 mice. **P < .01 by the nonparametric Mann-Whitney test. (D) qPCR analysis of hEBI2 transcript levels in spleen cells from young (12-15 weeks) and older, lymphoma IgH-hEBI2 mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 to 6 mice. ***P < .001 by the nonparametric Mann-Whitney test. (E-G) The B1a cell subsets in spleen (E), blood (F), and peritoneal cavity (G) samples from 4-day-old WT and IgH-hEBI2 mice were analyzed by FACS. The number of B1 B cells is given as percentage of total lymphocytes in the samples. The results are mean ± SEM of data from 3 to 7 mice. **P < .01 and ***P < .001 by Student t test.
Expression of EBI2 in lymphoma patients. (A) qPCR analysis of EBI2 transcript levels in CD10+ and CD10− cancers relative to the mean expression level of the healthy control samples. The cDNA samples in the array had been prenormalized to β-actin. The results are mean ± SEM of data from 6 to 14 samples. *P < .05 and ***P < .001 by Student t test. (B) Expression data on EBI2 expression in CLL patients compared with healthy controls. Expression data from Gene Expression Omnibus (GEO) profiles reprinted from Gutierrez et al,45 Gutiérrez et al46 with permission, and Fält et al47 with permission. (C) qPCR analysis of mEBI2 transcript levels in spleen cells from young (12-15 weeks) and older, lymphoma IgH-hEBI2 mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 to 6 mice. **P < .01 by the nonparametric Mann-Whitney test. (D) qPCR analysis of hEBI2 transcript levels in spleen cells from young (12-15 weeks) and older, lymphoma IgH-hEBI2 mice relative to the expression of the control gene GAPDH. The results are mean ± SEM of data from 5 to 6 mice. ***P < .001 by the nonparametric Mann-Whitney test. (E-G) The B1a cell subsets in spleen (E), blood (F), and peritoneal cavity (G) samples from 4-day-old WT and IgH-hEBI2 mice were analyzed by FACS. The number of B1 B cells is given as percentage of total lymphocytes in the samples. The results are mean ± SEM of data from 3 to 7 mice. **P < .01 and ***P < .001 by Student t test.
To study the relation between CLL and EBI2, we analyzed publicly available expression data from 2 studies in which CLL patients were compared with healthy controls. These studies strongly suggest that EBI2 is downregulated in human CLL (Figure 7B) cancers as compared with normal lymphatic tissues. In addition, progressive CLL samples seem to express even lover levels of EBI2 compared with stable, nonprogressive CLL samples (Figure 7B). Strikingly, these clinical expression patterns of EBI2 in CLL samples are accurately mimicked in IgH-hEBI2 mice where not only the transgenic hEBI2, but also the endogenous murine EBI2 (mEBI2) is highly downregulated in older mice with lymphomas compared with young adult (12- to 15-week-old) IgH-hEBI2 mice (Figure 7C-D). Human CLL has a very prolonged latency phase and the origin of the incurable CLL clones is obscure apart from the B1-like phenotype. B1 cells are typically generated during fetal development4 and we can show that the IgH-hEBI2 transgene can promote B1 cell expansion as early as 4 days after birth (Figure 7E-G). Together, these data indicate that although high expression of EBI2 in mice leads to an early expansion of highly proliferating B1 B cells, there is a strong negative selection on EBI2 for lymphoid cancer development, a phenomenon that may be mimicked during human CLL development.
Discussion
In this study, we establish that high expression of hEBI2 in B cells leads to a shift in B-cell subset distribution toward CD19+/B220low/− CD5+ B1a B cells. In combination with a hEBI2-induced increase in proliferation and antiapoptotic effects, this appears to predispose IgH-hEBI2 mice to develop CLL-like disease and die prematurely.
These findings add to the increasing knowledge we have about EBI2 as an important mediator of both innate and adaptive immune cell migration.5,6,34-37 The expansion of B1a cells observed in the spleen in this study could be due to proliferation and spillover of already existing B1a cells in the peritoneum, as we see a prominent expansion of B1 B cells in this compartment. Interestingly, findings similar to ours with an early observable expansion of B1 B cells in the peritoneum followed by expansion of the same B-cell subset in blood and spleen have been observed in 2 murine CLL models, namely Eμ-TCL1 mice38 and NZB IRF4+/− mice.39
Expansion of CD5+ B1 cell clones occurs spontaneously in the peritoneal cavites of aging mice, meaning that early CLL development occurs naturally over time.40 Most likely, the B1 cells are susceptible to genetic aberrations due to their continuous replenishment and strong survival signals provided through CD5,41 and these genetic aberrations may then occasionally lead to transformation events. Mice with an increased population of B1a cells from a young age, as is the case in our IgH-hEBI2 mice, will be even more vulnerable to such transformations. A central question is whether the EBI2-driven B1a CLL cells observed in our model are related to the CLL cells found in human cancers. Apart from the shared expression of CD5, the cellular origin of CLL in humans is still unknown, and the human counterpart of the murine B1 cells has not been found. Griffin et al have described a population of CD20+CD27+CD43+CD70− B cells, which are functionally similar to murine B1 B cells,42 but Covens et al later proposed that cells with this phenotype were more likely to correspond to preplasmablasts.43 Although the existence of B1 cells in humans is still being debated, a recent study used primates to identify B-cell populations from the peritoneal cavity, and here cells were identified, which were phenotypically and functionally similar to B1 cells from mice.44 This study hints to the fact that the B1 cell lineage and its functions and pathologies are conserved among mice and primates. The corresponding samples are not available to prove that human B1a cells exist in the peritoneal cavity of healthy individuals, but the observations in Eμ-TCL1 and our IgH-hEBI2 mice support the interpretation that murine CLL-like disease occurs when B1a cells expand and gradually dominate the lymphoid compartments. In this study, we have also provided evidence that the B1 B-cell expansion and thereby the cellular origin for the CLL-like cancer observed in IgH-hEBI2 mice is present immediately after birth and that disease progression is associated with a remarkable downregulation of both endogenous and transgenic EBI2 expression. As such, our findings may suggest that the phenotype of human CLL-causing cells is highly altered in secondary lymphoid organs as compared with the body cavities where the cells may initially accumulate to the highest degree. CLL currently stands as incurable despite the availability of effective firstline therapies often capable of achieving complete clinical remission. Our findings highlight the possibility of a CLL precursor with a markedly different phenotype to that of the clinically recognized cells. A search for such a precursor is not possible in our mice with polyclonal B1 cell expansion, but could be possible in future human studies where a particular gene signature could be searched for in patients during clinical remission.
In conclusion, we have demonstrated that EBI2 overexpression leads to expansion of a B1a B-cell subset, reduced immune response, upregulation of cellular oncogenes, late-onset lymphoid cancer development, late EBI2 downregulation, and premature death. These findings are highly similar to those observed in CLL patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Maibritt Sigvardt Baggesen and Randi Thøgersen (Department of Neuroscience and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark), Pernille Frederiksen (Department of Clinical Medicine, Rigshospitalet, Copenhagen, Denmark), and Heidi Marie Paulsen and Lise Schorling Strange (Department of Biomedicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark) for excellent technical assistance. The authors also thank Andreas W. Sailer (Novartis Institutes for BioMedical Research) for helpful suggestions in the project planning as well as comments on the manuscript.
This work was supported in part by the NovoNordisk Foundation, the Danish Council for Independent Research [Medical Sciences], the Lundbeck Foundation, the University of Copenhagen, the Aase and Einar Danielsen Foundation, the A. P. Møller Foundation for the Advancement of Medical Science (K.N.A., L.B., T.B.-J., V.K., V.D., K.S., P.J.H., and M.M.R.), and the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (A.L.K. and H.C.M.). V.K. was further supported by the Slovenian Research Agency (P4-0053).
Authorship
Contribution: K.N.A., L.B., T.B.-J., A.L.K., H.W., H.C.M., P.J.H., and M.M.R. conceptualized the study; K.N.A., L.B., T.B.-J., A.L.K., J.P.C., A.R.T., K.L.E., M.R.B., H.W., H.C.M., P.J.H., and M.M.R. developed the study methodology; K.N.A., L.B., T.B.-J., V.K., A.L.K., V.D., K.S., and P.J.H. conducted investigation; K.N.A., T.B.-J., P.J.H., and M.M.R. wrote the original draft; K.N.A., L.B., T.B.-J., A.L.K., H.C.M., P.J.H., and M.M.R. wrote, reviewed, and edited the manuscript; K.N.A., L.B., A.L.K., J.P.C., A.R.T., T.W.S., H.C.M., and M.M.R. acquired funding; and P.J.H. and M.M.R. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.B.-J. is Synaptic Transmission In Vitro, Lundbeck, Valby, Denmark.
The current affiliation for V.K. is Institute of Anatomy, Histology & Embryology, Veterinary Faculty, University of Ljubljana, Ljubljana, Slovenia.
Correspondence: Peter J. Holst, Center for Medical Parasitology, CSS, Building 22&23, Øster Farigmagsgade 5, DK-1014 Copenhagen, Denmark; e-mail: pholst@sund.ku.dk; and Mette M. Rosenkilde, Department of Neuroscience and Pharmacology, University of Copenhagen, Blegdamsvej 3b, Building 18.5, DK-2200 Copenhagen, Denmark; e-mail: rosenkilde@sund.ku.dk.