Key Points
TNFRSF14 and MAP2K1 mutations are frequent in PTFL but do not occur together in the majority of cases.
MAP2K1 mutations lead to activation of the downstream target phosphorylated extracellular signal-regulated kinase.
Abstract
Pediatric-type follicular lymphoma (PTFL) is a B-cell lymphoma with distinctive clinicopathological features. Recently, recurrent genetic alterations of potential importance for its pathogenesis that disrupt pathways associated with the germinal center reaction (TNFRSF14, IRF8), immune escape (TNFRSF14), and anti-apoptosis (MAP2K1) have been described. In an attempt to shed more light onto the pathogenesis of PTFL, an integrative analysis of these mutations was undertaken in a large cohort of 43 cases previously characterized by targeted next-generation sequencing and copy number array. Mutations in MAP2K1 were found in 49% (20/41) of the cases, second in frequency to TNFRSF14 alterations (22/41; 54%), and all together were present in 81% of the cases. Immunohistochemical analysis of the MAP2K1 downstream target extracellular signal-regulated kinase demonstrated its phosphorylation in the evaluable cases and revealed a good correlation with the allelic frequency of the MAP2K1 mutation. The IRF8 p.K66R mutation was present in 15% (6/39) of the cases and was concomitant with TNFRSF14 mutations in 4 cases. This hot spot seems to be highly characteristic for PTFL. In conclusion, TNFRSF14 and MAP2K1 mutations are the most frequent genetic alterations found in PTFL and occur independently in most cases, suggesting that both mutations might play an important role in PTFL lymphomagenesis.
Introduction
Pediatric-type follicular lymphoma (PTFL) has been recognized as a definitive entity in the revised 2016 World Health Organization Lymphoma Classification.1,2 Until recently, little was known about the genetic alterations involved in the pathogenesis of this disease. Nevertheless, several groups have described a specific mutational profile in PTFL distinct from other non-Hodgkin lymphomas, including conventional follicular lymphoma (FL), by using next-generation sequencing (NGS) technologies and copy number (CN) arrays.3-5 Overall, PTFL lacks mutations of histone modifying genes frequently found in FL and shows low levels of genomic complexity in concordance with its indolent clinical behavior. The most frequently mutated genes reported in PTFL are TNFRSF14, MAP2K1, and IRF8, albeit at different frequencies in published series.3-6 Furthermore, aberrations including CN neutral loss of heterozygosity (CNN-LOH) of the 1p36 region, containing TNFRSF14, have also been found in PTFL, suggesting a tumor suppressor function of this gene in this disease.3,5-7
Mutations observed in PTFL are not restricted to this disease and are well known to occur in other types of non-Hodgkin lymphoma. Mutations in TNFRSF14 occur at high frequencies also in adult FL (18% to 44%)8 and diffuse large B-cell lymphoma (DLBCL, 22%).9 MAP2K1 mutations have been described as driver mutations in hairy-cell leukemia variant (HCLv) and/or conventional HCL with immunoglobulin heavy-chain V4-34+,10,11 Langerhans cell histiocytosis,12-14 and in isolated cases of chronic lymphocytic leukemia (CLL),15,16 splenic marginal zone lymphoma,17,18 and splenic diffuse red pulp lymphoma.19 IRF8 gene mutations have also been identified in DLBCL and FL, but without apparent functional consequences.9,20-22
A drawback in understanding the genetic landscape of PTFL is that the occurrence of these alterations has been described in different, mostly small cohorts of PTFL cases, and the cooccurrence of these mutations and their relevance for PTFL lymphomagenesis is not known. By expanding the genetic analysis of our published PTFL cohort5 for MAP2K1 and IRF8 mutations, and performing an integrative analysis, we wanted to clarify their frequency and overlap with TNFRSF14 mutations.
Study design
Cases
A total of 43 well-characterized PTFL cases were included. Clinical and morphological features were previously reported.5
Mutational analysis
All cases have been genetically characterized by targeted NGS and CN arrays using formalin-fixed paraffin-embedded tissue. Forty-one PTFL cases were additionally investigated for MAP2K1 mutations using a single-amplicon NGS approach covering exons 2 and 3. The amplicons were analyzed on the Ion Torrent PGM (Thermo Fisher Scientific, Schwerte, Germany), as previously described.5 The mean coverage of the amplicons was 18 132 reads (range, 104-79 524 reads). Sanger sequencing was performed to detect the IRF8 p.K66R variant in 39 PTFL using primers previously described.4 Allelic frequencies of these mutations are in the range of Sanger sequencing detection.4 Sequence analysis was performed using Mutation Surveyor software (SoftGenetics LLC, State College, PA). The mutational analysis results were integrated to the previous genetic analysis.
Immunohistochemical analysis
Immunohistochemical analysis of phosphorylated extracellular signal-regulated kinase protein (pERK) (Cell Signaling Technologies) was performed in formalin-fixed paraffin-embedded sections in an automated immunostainer (Ventana Medical System, Tucson, AZ).
Results and discussion
Twenty of 41 PTFL (49%) cases carried MAP2K1 mutations (Figure 1A). MAP2K1 mutations were identified mainly in 2 hot spots within exon 2 (codons 53 and 57), which encode the negative regulatory region domain of MEK1 protein, corroborating previous results in PTFL,3 HCLv,10,11 and CLL.15,16 In contrast, mutations in Langerhans cell histiocytosis spread across exons 2 and 3.12-14 The allelic frequencies of MAP2K1 mutations ranged between 4% and 35% (median, 10%). The frequency of MAP2K1 mutations identified is similar to what we reported for TNFRSF14 (21/41; 51%).5 MAP2K1 and/or TNFRSF14 mutations were observed in 33/41 cases (81%); however, only 8 cases (8/41; 20%) showed mutations in both genes, whereas the majority of cases had either a TNFRSF14 (13/41; 32%) or a MAP2K1 (12/41; 29%) mutation. This finding indicates that both genes independently are of importance for the pathogenesis of PTFL, despite their different functional properties. TNFRSF14 mutations abrogate the interaction between TNFRSF14 and BTLA (B and T lymphocyte attenuator) receptors disrupting an important tumor suppressor axis that leads to B-cell receptor activation.7 The frequent TNFRSF14 mutations together with CNN-LOH of 1p36 indicate a powerful selection against the TNFRSF14 gene during PTFL development.
Distribution of MAP2K1 mutations at protein and exon level in PTFL in comparison with other hematological neoplasias. (A) Schematic diagram of MAP2K1 mutations in PTFL,3,4 Langerhans cell histiocytosis (LCH),12-14 hairy cell leukemia10,11 (including HCLv and conventional HCL [HCLc] with IGHV4-34+), splenic diffuse red pulp small B-cell lymphoma (SDRPL),19 splenic marginal zone lymphoma (SMZL),17,18 CLL,15,16 according to NGS studies and/or Sanger analysis. Exons are represented by boxes on the body of MEK1 protein and the main protein domains are represented by larger colored boxes. AL, activation loop; DD, docking domain for ERK1 and ERK2; DVD, domain of versatile docking (MAP3K docking domain); NES, nuclear export sequence; NRR, negative regulatory region; PRD: proline-rich domain. (B) Immunohistochemical analysis of pERK in MAP2K1 mutated and wild-type PTFL cases. Note that the variant allelic frequency (VAF, indicated in parentheses) of MAP2K1 mutations by NGS analysis correlates with the amount of pERK-positive cells.
Distribution of MAP2K1 mutations at protein and exon level in PTFL in comparison with other hematological neoplasias. (A) Schematic diagram of MAP2K1 mutations in PTFL,3,4 Langerhans cell histiocytosis (LCH),12-14 hairy cell leukemia10,11 (including HCLv and conventional HCL [HCLc] with IGHV4-34+), splenic diffuse red pulp small B-cell lymphoma (SDRPL),19 splenic marginal zone lymphoma (SMZL),17,18 CLL,15,16 according to NGS studies and/or Sanger analysis. Exons are represented by boxes on the body of MEK1 protein and the main protein domains are represented by larger colored boxes. AL, activation loop; DD, docking domain for ERK1 and ERK2; DVD, domain of versatile docking (MAP3K docking domain); NES, nuclear export sequence; NRR, negative regulatory region; PRD: proline-rich domain. (B) Immunohistochemical analysis of pERK in MAP2K1 mutated and wild-type PTFL cases. Note that the variant allelic frequency (VAF, indicated in parentheses) of MAP2K1 mutations by NGS analysis correlates with the amount of pERK-positive cells.
Because MAP2K1 mutations are predicted to constitutively activate the downstream ERK1/2 proteins by phosphorylation, 12 PTFL cases (6 MAP2K1 mutated cases and 6 wild-type cases), 3 reactive lymph nodes, and 3 tonsils were analyzed with a pERK antibody by immunohistochemistry.12-14 In normal lymph nodes, tonsils, and PTFL cases without MAP2K1 mutation, the staining was negative in the germinal center (GC) cells with positive internal control in the endothelial cells (Figure 1B). The analysis in the PTFL cases revealed a good correlation of pERK staining and allelic frequency of MAP2K1 mutation in the GC cells in the different cases, and confirmed the downstream activation of ERK in PTFL (Figure 1B; supplemental Table 1, available on the Blood Web site). Despite different functions, both genes/proteins have in common the control of signaling pathways important for B-cell proliferation.
Another interesting finding is the higher allelic frequency of TNFRSF14 mutations in comparison with MAP2K1 mutations, including 5 cases with both mutations (median ± standard deviation, 17.8 ± 6.3, vs 10 ± 8.6, P = .046), even after correcting for concomitant CNN-LOH of 1p36 found in 14/20 TNFRSF14-mutated cases. This suggests that TNFRSF14 mutations occur earlier in tumorigenesis than MAP2K1 mutations (supplemental Figure 1).
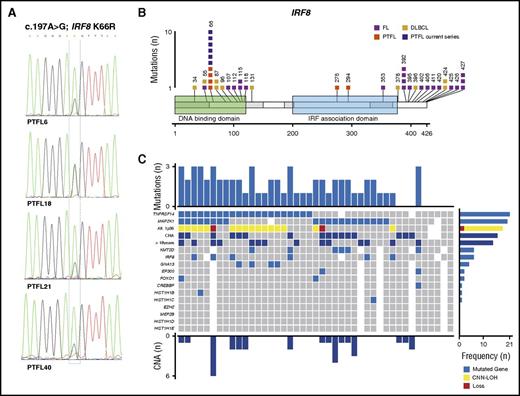
The IRF8 mutations at the hotspot p.K66R (c.197A>G), affecting the DNA binding domain, were recently reported to be specific for PTFL.4 Accordingly, 6 of 39 cases analyzed (15%) carried this mutation (Figure 2A), and only 1 case (PTFL18) showed a concomitant 16q CNN-LOH alteration. The frequency of IRF8 mutations found in the present study was lower than that described by Ozawa et al (15 vs 50%).4 The difference might be explained by the small cohort of 6 cases analyzed in that study. The IRF8 gene has been described as potential tumor suppressor in myeloid neoplasms and has recently also been linked to the pathogenesis of B-cell lymphomas. Specifically, IRF8 mutations have been observed in adult FL and DLBCL in 5% to 10% of cases.9,20,21 However, in contrast to PTFL where the IRF8 p.K66R mutation is predicted to affect DNA-protein interaction,4 these mutations are frequently indel and missense mutations predominantly located in the IRF8 C-terminal domain (Figure 2B) with still unidentified functional consequences.9,20-22 Of note, IRF8 and TNFRSF14 are critical regulators of the immune system development and function.7,23 Both genes regulate GC B-cell activation,7,23 and deficiency of IRF8 has been reported to induce a hyperproliferative phenotype in pre-B cells.24 Interestingly, 4 of 6 IRF8 mutated cases also showed TNFRSF14 mutations, suggesting possible cooperation between these 2 genes.
IRF8 mutations and global mutational landscape in PTFL. (A) Sequence electropherograms from cases PTFL6, PTFL18, PTFL21, and PTFL40 showing IRF8 p.K66R (c.197A>G) mutation by Sanger sequencing. PTFL18 carried a concomitant 16q11.2-q24.3 CNN-LOH, including an IRF8 gene. (B) Schematic diagram of IRF8 mutations in FL,20-22 DLBCL,9,25 and PTFL,3,4 according to NGS studies. Exons are represented by boxes on the body of the protein and the main protein domains are represented by larger colored boxes. Domains of the protein are represented according to Uniprot database (www.uniprot.org).4 (C) Overview of the global mutational landscape in 43 PTFL cases. Each column of the heat map represents 1 PTFL case and each line 1 specific analysis. On the right side of the figure, the frequency of the particular result of the analysis is shown.
IRF8 mutations and global mutational landscape in PTFL. (A) Sequence electropherograms from cases PTFL6, PTFL18, PTFL21, and PTFL40 showing IRF8 p.K66R (c.197A>G) mutation by Sanger sequencing. PTFL18 carried a concomitant 16q11.2-q24.3 CNN-LOH, including an IRF8 gene. (B) Schematic diagram of IRF8 mutations in FL,20-22 DLBCL,9,25 and PTFL,3,4 according to NGS studies. Exons are represented by boxes on the body of the protein and the main protein domains are represented by larger colored boxes. Domains of the protein are represented according to Uniprot database (www.uniprot.org).4 (C) Overview of the global mutational landscape in 43 PTFL cases. Each column of the heat map represents 1 PTFL case and each line 1 specific analysis. On the right side of the figure, the frequency of the particular result of the analysis is shown.
In conclusion, this integrative genetic analysis demonstrated that in 88% of PTFL cases (38/43), 1 or several alterations/genetic events were identified (Figure 2C). Only 5 cases (12%) did not show genetic alterations other than immunoglobulin heavy-chain rearrangement. TNFRSF14 and MAP2K1 mutations are found in about half of the cases each, but do not occur together in the majority of the cases, indicating important but distinct roles in lymphomagenesis for both genes. We confirmed that IRF8 p.K66R mutations are highly specific for PTFL but occur less commonly than other events. Nevertheless, the similar functions and often cooccurrence of IRF8 and TNFRSF14 mutations suggest that these 2 mutations might cooperate in the pathogenesis of PTFL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Noelia Garcia, Silvia Martín, Sieglinde Baisch, Claudia Hermann, and Sema Colak for excellent technical assistance.
This work was partially developed at the Centro Esther Koplowitz, Barcelona, Spain. This work was supported by Fondo de Investigaciones Sanitarias Instituto de Salud Carlos III (CP13/00159 and PI15/00580) (I.S.), Generalitat de Catalunya Suport Grups de Recerca (2013-SGR-378 I.S. and 2014-SGR-795) (E.C.), the European Regional Development Fund “Una manera de fer Europa” and the Wilhelm-Sander-Stiftung (2015.058.1) (L.Q.-M. and F.F.). J.S. was supported by the Wilhelm-Sander-Stiftung. J.E.R.-Z. was supported by a fellowship from Generalitat de Catalunya AGAUR FI-DGR 2017 (2017 FI_B01004). E.C. is an Academia Researcher of the “Institució Catalana de Recerca i Estudis Avançats” of the Generalitat de Catalunya.
Authorship
Contribution: L.Q.-M. and I.S. conceived and designed the study, supervised the experimental work, and wrote the manuscript; J.S., I.B., J.E.R.-Z., A.N., and I.S. performed genetic analysis and interpreted the data; F.N. performed bioinformatics analysis; B.G.-F., T.M., J.C., J.v.d.W., A.R., G.O., S.D., C.E., E.S.J., F.F., E.C., and L.Q.-M. contributed with cases; I.A.M.-M. and L.Q.-M. performed and interpreted the immunohistochemical analysis; and J.S., E.S.J., E.C., and F.F. analyzed the data and helped writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Leticia Quintanilla-Martinez, Institute of Pathology, Tübingen University Hospital, Liebermeisterstr 8, 72076 Tübingen, Germany; e-mail: leticia.quintanilla-fend@med.uni-tuebingen.de; and Itziar Salaverria, Institut d’Investigacions Biomèdiques August Pi i Sunyer, Rosselló 153, Barcelona, 08036, Spain; e-mail: isalaver@clinic.ub.es.
References
Author notes
J.S. and J.E.R.-Z. are joint first authors.
E.S.J., I.S., and L.Q.-M. are joint senior authors.

![Figure 1. Distribution of MAP2K1 mutations at protein and exon level in PTFL in comparison with other hematological neoplasias. (A) Schematic diagram of MAP2K1 mutations in PTFL,3,4 Langerhans cell histiocytosis (LCH),12-14 hairy cell leukemia10,11 (including HCLv and conventional HCL [HCLc] with IGHV4-34+), splenic diffuse red pulp small B-cell lymphoma (SDRPL),19 splenic marginal zone lymphoma (SMZL),17,18 CLL,15,16 according to NGS studies and/or Sanger analysis. Exons are represented by boxes on the body of MEK1 protein and the main protein domains are represented by larger colored boxes. AL, activation loop; DD, docking domain for ERK1 and ERK2; DVD, domain of versatile docking (MAP3K docking domain); NES, nuclear export sequence; NRR, negative regulatory region; PRD: proline-rich domain. (B) Immunohistochemical analysis of pERK in MAP2K1 mutated and wild-type PTFL cases. Note that the variant allelic frequency (VAF, indicated in parentheses) of MAP2K1 mutations by NGS analysis correlates with the amount of pERK-positive cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/3/10.1182_blood-2017-03-776278/4/m_blood776278f1.jpeg?Expires=1765082802&Signature=mFxgXRESXwzxA5OoGyA37M4p9wn7TDlU6FFiqPRnvPxYm7soWRUPIujBu2GzozibyNpr9V2quFi3DpGjqxlOg30Md~Xg0InCBeFRD2yTFg42~S2BPh0XPa6BYW3RMCuEfUEE0-TNZA9GCIp247BHp26Mo42zuv~BvlbIp~vIpE48rf3KDPKelh47wXJCqYQ8f28~Mfg5ml069TxvvL7f-cMGFnVJ3fwszwAxZS~qBNHmUTwxTzopZWSrwzPZ1WvKYz2OS8lRywJHJbMe2aXaJRq1ydCGNlXKaBhLsKuGFdDjjczTliKenKFnBctFK1H~VVFVas2uOH28nmGFmmWSDw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)