In this issue of Blood, Hu et al demonstrate that homozygous factor 10 gene ablation (f10−/−) in the zebrafish does not result in either embryonic lethality or vascular developmental abnormalities.1
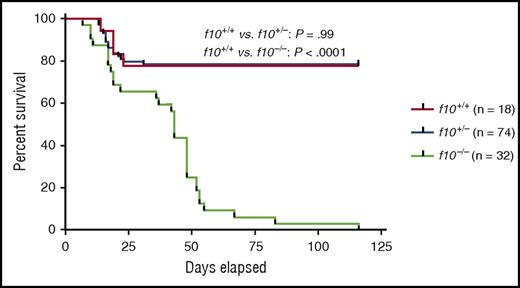
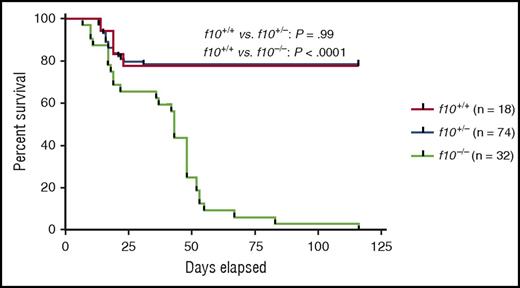
Survival curves of closely monitored clutches of zebrafish offspring from f10+/− incrosses demonstrate progressive loss of 75% of homozygotes by 50 dpf and 100% by 115 dpf. There was no significant loss of heterozygotes (P > .05 by log-rank testing). Larvae were genotyped at 3 dpf, and selected individuals were observed daily. There was ∼20% background loss of individual fish across all genotypes up to 20 dpf, which is typical during wild-type fish development. See Figure 3B in the article by Hu et al that begins on page 666.
Survival curves of closely monitored clutches of zebrafish offspring from f10+/− incrosses demonstrate progressive loss of 75% of homozygotes by 50 dpf and 100% by 115 dpf. There was no significant loss of heterozygotes (P > .05 by log-rank testing). Larvae were genotyped at 3 dpf, and selected individuals were observed daily. There was ∼20% background loss of individual fish across all genotypes up to 20 dpf, which is typical during wild-type fish development. See Figure 3B in the article by Hu et al that begins on page 666.
The f10−/− phenotype maintains the expected Mendelian ratios during early development, before exhibiting delayed mortality between 16 and 120 days postfertilization (dpf) as a result of spontaneous bleeding (see figure). These results contrast with murine gene ablation studies for the common pathway coagulation factors (F10, F5, and prothrombin), which demonstrate a mixture of embryonic lethality and fatal perinatal bleeding.2-5
F10 deficiency in humans is a rare autosomal recessive bleeding disorder in which the majority of reported genetic defects are either homozygous or compound heterozygous point mutations (http://www.omim.org/entry/613872). The bleeding phenotype depends on the residual coagulant function, with severe cases possessing <1% activity. F10 gene ablation in the mouse demonstrates partial embryonic lethality with approximately one-third of the f10−/− embryos dying from day 11.5 to 12.5. The remainder survive to term but bleed to death shortly thereafter (90% within 5 days of birth).2 Similarly, murine f5 and prothrombin gene ablations demonstrate partial embryonic lethality between days 9.5 and 10.5 and are associated with yolk sac vascular anomalies. The remainder of the embryos seem to succumb to a hemorrhagic death in late gestation or shortly after birth.3-5 The phenotypic similarities among these common coagulation pathway gene defects, along with similar partial embryonic lethality in the protease-activated receptor-1 (PAR-1) gene ablation,6 led to the hypothesis that thrombin-mediated proteolysis is crucial for vascular development.
Hu et al now clearly demonstrate that the f10−/− genotype is compatible with normal embryonic vertebrate development in the zebrafish. Furthermore, detailed comparison of vasculogenic marker expression and vessel anatomy failed to demonstrate significant differences between wild-type and f10−/− zebrafish. The f10−/− genotype exhibited delayed mortality (16-120 dpf) associated with predominantly brain, muscle, and intra-abdominal bleeding. Pooled citrated plasma samples from 1-month-old f10−/− fish did not generate detectable thrombin activity (by human fibrinogen clotting) whereas plasma from the f10+/+ genotype developed activity within minutes. Hemostasis was defective from early development (3 dpf) with a markedly prolonged time to occlusion in response to laser-induced endothelial injury. This defect could be rescued by f10 transgene injection but not by an anti-fibrinolytic agent. Finally, as proof of principle, transgene injection was used in the laser injury model to test the hemostatic function of human F10 variants of unknown significance by engineering these mutations into orthologous positions in the zebrafish f10 complementary DNA.
Murine studies (and mammalian studies in general) are complicated by the potential maternal contribution to coagulation factor deficiencies. The murine F10 Friuli knockin model (1% to 3% activity) demonstrates that minimal amounts of functional zymogen are sufficient to rescue embryonic/perinatal lethality, with older mice showing colocalized iron deposition and cardiac fibrosis from recurrent hemorrhage.7 In contrast, the maternal contribution to zebrafish f10 expression was no longer detectable by 3 dpf, allowing a more straightforward interpretation of the zebrafish phenotype. In addition, genome editing technologies including transcription activator-like effector nucleases (TALENs) and clustered regularly interspaced short palindromic repeats and associated Cas genes (CRISPR/Cas9) now allow definitive genetic approaches in the zebrafish, as opposed to gene knockdowns with variable residual expression.
What are the differences between the murine and zebrafish systems that result in these phenotypic disparities? One possibility is that species-specific differences exist between the zebrafish and higher vertebrate hemostatic systems. The zebrafish genome includes the teleost-specific whole-genome duplication,8 which generated paralogues of numerous genes including coagulation proteins. Subfunctionalization of paralogues into distinct hemostasis and development roles would potentially obscure their mammalian function(s). Similarly, genetic background has profound effects on gene ablation phenotypes in inbred mouse strains, but these effects are less discernible in hybrid zebrafish backgrounds. Finally, phenotypic differences may reflect the developmental milieu: yolk-based embryonic development in an aquatic environment vs placental-based development and birthing in a terrestrial environment. Although the underlying hemophilic tendencies may be similar, the hemostatic challenges seem to be more frequent and profound in the terrestrial environment, given the dependence on placental feto-maternal exchange and relative trauma of the birthing process. Indeed, mortality in the f10−/− zebrafish accelerates at 1 to 2 months after fertilization, correlating with the transition to sexual maturity and increase in aggressive behaviors. This observation parallels the role that hemostatic challenges can play in the recognition of clinical bleeding disorders.
The lack of embryonic lethality in the zebrafish f10−/− genotype provides additional opportunities to identify genetic modifiers of this phenotype, both known and unknown. This approach plays to the strengths of the zebrafish model and may identify novel approaches to rebalancing hemostasis in the treatment of bleeding disorders. In addition, although the zebrafish f10−/− phenotype does not support a role for thrombin-mediated proteolysis in vasculogenesis, alternative pathways for thrombin generation during zebrafish development are not yet formally excluded. With the recent progress in genome editing technologies, the zebrafish model continues to become an even more powerful tool for investigating the genetic basis of bleeding and thrombotic disorders.
Conflict-of-interest disclosure: J.P.S. has received research funding from the Bayer Hemophilia Awards Program and the Pfizer ASPIRE Hemophilia program and performed grant review for the latter program.