To the editor:
Acute vaso-occlusive pain episodes are a hallmark of sickle cell anemia (SCA), one of the most common Mendelian disorders worldwide with an estimated >300 000 births annually.1-3 Although SCA is a monogenic disorder, manifestations and disease severity are highly variable, suggesting additional phenotypic modifiers. The few genetic factors known to act as phenotypic modifiers do not completely explain the clinical heterogeneity in SCA. Previous genetic association studies identified that variants at 3 distinct loci (BCL11A, HBS1L-MYB, and HBB) are strong determinants of fetal hemoglobin level, and the single-nucleotide polymorphism (SNP) variant rs6141803 located upstream of COMMD7 is associated with acute chest syndrome.4 Also, heme oxygenase-1 gene promoter polymorphisms influence heme oxygenase (HO-1) activity and the incidence of acute chest syndrome in children with sickle cell disease (SCD).5,6 We conducted this genome-wide association study (GWAS) to identify the variants associated with acute, severe vaso-occlusive pain in children with SCA enrolled in the Cooperative Study for Sickle Cell Disease (CSSCD) and Silent Infarct Transfusion (SIT) trial.
The CSSCD, a multi-institutional prospective cohort, natural history study of SCD, enrolled 3538 individuals with SCD between 1979 and 1981.7 The SIT trial, a multicenter international trial, screened 1210 children with SCA to test the hypothesis that regular blood transfusions attenuate progression of cerebral infarcts in children with preexisting silent strokes.8 Both studies were approved by the Institutional Review Boards at Boston University School of Medicine and Vanderbilt University Medical Center.
We included participants identified as being of African descent from both cohorts with available genotype data, and who were diagnosed with SCA. We excluded participants if essential clinical or demographic data (necessary for phenotypic assignment or previously reported to impact the pain phenotype) were missing, or if there was discordance between genetically defined and self-identified sex. We excluded all self-reported first-degree relatives, and cryptic relatedness (including full siblings, parents, and offspring) determined by examining pairwise identity-by-descent in the combined cohort. To harmonize pain phenotypes in the CSSCD and SIT trial cohorts, the age inclusion criterion of 2 to 18 years was used to match the age and length of follow-up in both cohorts. SIT trial participants were between 5 and 15 years of age at the time of registration and the trial included a 3-year retrospective collection of all acute, severe vaso-occlusive pain based on hospitalization and treatment with opioid medication.8,9 Unlike previous CSSCD pain analyses, where the definition of a pain episode included an acute vaso-occlusive event that lasted at least 2 hours and resulted in a physician visit,9 we restricted the definition of a pain episode to include only episodes requiring hospitalization to match the SIT trial definition.
CSSCD cohort DNA samples were genotyped at Boston University School of Medicine by using Illumina Human610-Quad arrays (n = 610 000 SNPs) (Illumina, San Diego, CA) and BeadStudio was used to call genotypes. SIT trial samples were genotyped at the Center for Inherited Disease Research at Johns Hopkins University School of Medicine (N = 573) by using the Illumina HumanHap650Y array (n = 661 000 SNPs) (Illumina) or at the Centers for Disease Control and Prevention (Atlanta, GA) (N = 509) by using the Illumina Infinium HumanOmni1-Quad array (n = 1 134 514 SNPs) (Illumina). After detailed quality control procedures were completed (and excluding CSSCD samples outside the SIT trial age inclusion criteria [≥2 and ≤12 years of age]), 359 and 934 samples from the CSSCD and SIT cohorts, respectively, were included in the analysis (Table 1; supplemental Figure 1A-B; and supplemental Methods, available on the Blood Web site). To infer ungenotyped SNPs and fill in missing data across genotyping platforms in the SIT trial and CSSCD cohort, we merged HumanHap650Y, HumanOmni1-Quad, and Human610-Quad array data sets and performed imputation for autosomal markers by using a Hidden Markov model as implemented in Markov Chain Haplotyping algorithm (MaCH), version 1.16,10 with 50 rounds and 200 states. Quality control was performed both before and after imputation and poorly imputed SNPs (squared correlation between imputed and true genotypes < 0.3) were excluded; a total of 1 098 907 SNPs remained for analysis. Due to the observed overdispersion of pain episodes in both cohorts, a multivariate quasi-Poisson regression model, with correction for estimated overdispersion, was used to evaluate the possible associations between SNPs and the acute severe vaso-occlusive pain rate, which was treated as a quantitative trait. The model was adjusted for age at enrollment, sex, hematocrit, and the top 10 principal components from the genetic data (to account for population substructure and genetic heterogeneity), assuming additive effects of allele dosage on the acute vaso-occlusive pain rate.
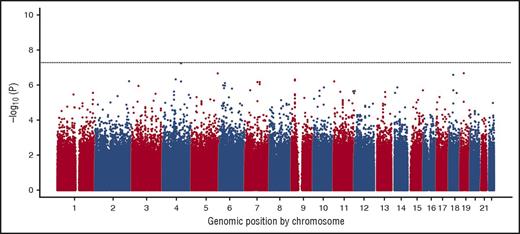
Participant characteristics for the SIT trial and CSSCD cohort are shown in Table 1. Statistically significant, but not clinically relevant, differences were identified between the 2 cohorts in age, percentage of fetal hemoglobin, reticulocyte percentage, pain rate, and follow-up time. The Manhattan plot summarizing the results of the GWAS for acute vaso-occlusive pain in the SIT trial and CSSCD cohort for the additive model is shown in Figure 1. The genomic inflation λ coefficient was 1.079, suggesting minimal test-statistic inflation by potential population stratification, cryptic relatedness, or other technical factors. Although none of the SNPs were significant at P < 5.0 × 10−8, 1 novel locus approached genome-wide significance: SNP rs3115229 (P = 5.63 × 10−8). This SNP is located 63.7 kb 5′ upstream of the KIAA1109 gene on chromosome 4 (4q27).
Manhattan plot showing the genome-wide –log10P values for association of SNPs with vaso-occlusive pain. Only 1 SNP on chromosome 4 (rs3115229) approached genome-wide significance (P = 5.63 × 10−8).
Manhattan plot showing the genome-wide –log10P values for association of SNPs with vaso-occlusive pain. Only 1 SNP on chromosome 4 (rs3115229) approached genome-wide significance (P = 5.63 × 10−8).
The suggested locus includes the KIAA1109-TENR-IL2-IL21 linkage disequilibrium block, containing 3 known protein-coding genes, TENR, IL2, and IL21, and a predicted gene of unknown function, KIAA1109. This locus has been associated with autoinflammatory disorders, such as celiac disease,11,12 ulcerative colitis,13,14 and rheumatoid arthritis.15,16
Given the nature of GWAS studies, namely associations between a SNP and a phenotype, we can only postulate as to the potential role of this locus in the pathogenesis of acute vaso-occlusive pain, a complex phenomenon involving tissue ischemia, hypoxia-reperfusion injury, immune responses and inflammation,17,18 and interactions between red blood cells, the endothelium, and leukocytes regulated by T-cell cytokines and adhesion molecules.19,20 Interleukin 2 (IL-2) and IL-21 may modulate acute pain in SCD through their effects on inflammation and immune responses. IL-2 is a key cytokine for T-cell activation and proliferation.21 IL-21 enhances B, T, and natural killer cell proliferation and interferon-γ production; inhibiting IL-21 has been shown to dampen inflammatory responses.22,23 T lymphocytes have also been implicated as mediators of pain hypersensitivity.24 KIAA1109 is moderately expressed in all adult and fetal tissues and encodes a protein of unknown function.25 TENR encodes testis nuclear RNA-binding protein, expressed primarily in the testis.
Strengths of the study include the consistent definition of acute, severe vaso-occlusive pain requiring hospitalization, the relatively large non–hydroxyurea treated population (a potential confounder) from 2 independent cohorts of children not on disease modifying therapy with hydroxyurea, with fewer comorbidities, and lower rates of chronic pain than adults with SCA. Pooling the cohorts improved power for our discovery analysis, but precluded validation in a separate cohort.
In summary, we present preliminary evidence of an association between variant rs3115229 and acute, severe vaso-occlusive pain in children with SCA. Our results will require additional validation and functional studies to understand the biology and reveal mechanisms by which candidate SNPs/genes might have their effects.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank the families and children with SCD who were participants in the SIT trial and CSSCD.
This work was supported by the National Institutes of Health (NIH) National Heart, Lung, and Blood Institute (U54HL090515, 4U01HL117721, 5R01HL091759, R01 HL87681 [M.H.S.], R01 HL 068970 [M.H.S.], and T32 HL007501 [J.N.M.]), and the NIH National Institute of Neurological Disorders and Stroke (5U01-NS042804-03); the Burroughs Wellcome Foundation (M.R.D.); an American Society of Hematology Research Training Award for Fellows (S.C.); and the Jim and Carol O’Hare Fellowship (S.C.).
The funders had no role in study design, data collection and analysis, preparation of the manuscript, or decision to publish. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Contribution: M.R.D. designed the research and wrote portions of the manuscript; P.B. performed analysis and interpreted the data; S.C. interpreted the data and wrote the manuscript; C.J.B., J.N.M., D.E.A., E.B.-C., J.F.C., and M.H.S. designed the study, collected data, interpreted the analysis, and wrote the manuscript; and all authors read and approved the final draft of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael R. DeBaun, Division of Hematology/Oncology, Vanderbilt-Meharry Center in Sickle Cell Disease, 2200 Children’s Way, Suite 11206, Nashville, TN 37232; e-mail: m.debaun@vanderbilt.edu.