Abstract
Background
Next generation sequencing (NGS) approaches have transformed our understanding of the genetic heterogeneity of Chronic lymhocytic leukemia (CLL). However, the detection of somatic mutations and their relative frequencies is variable, which possibly reflects differences in the composition of the cohorts studied worldwide and the time point during the course of the disease when they were tested.
Considering the landscape of genomic alterations recently identified in CLL as well as the availability of new targeted therapies, it is warranted to accurately predict outcome at the individual patient level. An international prognostic index (CLL-IPI) and the molecular index (M-IPI) are promising scoring systems to improve the precision of prognostic counseling.
Aim
The aims of the study were: 1) To characterize CLL patients not previously treated according to their biological profile and their impact on the prognosis; 2) To test the hierarchical of both stratification models in general practice.
Patients and methods
We included 205 patients consecutively diagnosed of CLL between 1986 and 2016 with available DNA at diagnosis. Clinical and biological data were extracted from medical records. IGVH mutational status were identify according to the updated ERIC recommendations. A custom panel designed by Sequencing Multiplex was used to investigate somatic mutations with MiSeq platform. It included the following genes; TP53 (all), BIRC3 (ex 7-9), SF3B1 (ex 14-16), MYD88 (ex 5), and NOTCH1 (ex 34), ATM (all), XPO1 (ex 15-16), EGR2 (ex 2), POT1 (ex 4-9) and NFKBIE (ex 1).
Bioinformatics analysis was achieved by Strand-NGS. The statistical analysis was completed with SPSS Version 20.0 and R Version 2.15.1 software. Times to first-treatment (TFT) curves were plotted using the Kaplan-Meier method and compared by the log-rank test.
Results
Median age was 64 years (range, 32-93) with a 1.2:1 ratio of males to female. The median follow-up was 76 (1-295) months. In our cohort, 166 (83%) patients were in Binet A, and 34 (17%) Binet B-C. Concerning Rai stage 123 (61,5%) were Rai 0 and 77 (38,5%) Rai I-IV. In 118 out of 203 (58,1%) IGVH was germline.
The most frequent cytogenetic abnormalities was del(13q) in 61 (32,1%), following +12 in 39 (10,5%), del (17p) in 11 (5,9%) and del(11q) in 8 (4,2%) and 17 missed data.
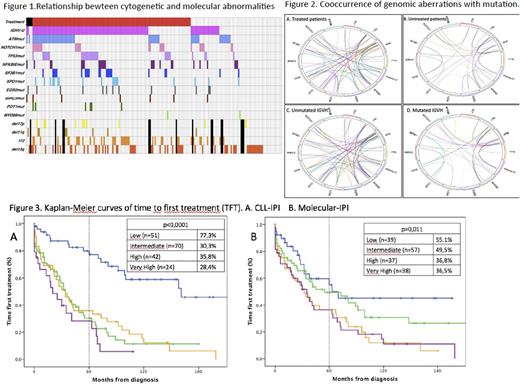
Regarding the molecular data, 61 (29,8%) carried ATM mutation, 30 (14,6%) harbored a TP53 mutation, 26 (12,7%) NOTCH1, 24 (11,7%) NFKBIE, 22 (107 (3,4%) BIRC3, 6 (2,9%) POT1 and 2 (1%) carried a MYD88 hotspot p.L265P mutation. Among the 40 patients with TP53 disruption, 29 had TP53 mutations, 11 had only del(17p) and 6 had both of them. Frequencies and distributions of mutations, genomic aberrations, IGHV status and need of treatment are illustrated in Figure 1.
At least 1 mutation was identified in 109 of 171 (63,7%) patients; 60 (35,1%) patients had 1, 34 (19,9%) had 2, 13 (7,6%) had 3, and two cases had 4 and 5 mutations respectively. Cooccurrence of genomic aberrations with mutations for all patients are shown in Figure 2.
Most frequent chemotherapy schedules were: Fludarabine-cyclophosphamide-rituximab (FCR) 39 (36,1%), Chlorambucil 36 (33,3%), bendamustine-rituximab 26 (24,1%), Obinutuzumab-Rituximab 4 (2%) and Ibrutinib 3 (1,5%). Five patients transformed into Richter syndrome.
Presence of TP53 mutations significantly correlated with unmutated IGVH, increased β2-microglobulin levels, CD38 expression, Richter's Syndrome development and treatment requirement. ATM mutations associated with unmutated IGVH, LDH increased levels and del(11q). Either, IPI-CLL and molecular-IPI identified 4 groups with significantly different outcome in terms of TFT although this difference was marginal in the high and very high categories. The percentage of patients in each group identified by the two scores as well as the TFT is detailed in Figure 3. In our series, IPI-CLL discriminated better than molecular-IPI, (p<0,0001 and 0.011, respectively).
Conclusions
In our cohort, gene mutations frequencies were very similar to those previously described in the literature. Our study highlights that both CLL-IPI and M-IPI are useful tools for real-life practice as both identified four group of patients with significantly different TFT times.
Samples provided by INCLIVA Biobank (PT13/0010/0004). Study supported by: FISS PI 10/02095, FISS PI 14/02018, FEHH (Spanish Hematologic Fundation)
Solano: Neovii: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.