Abstract
Background: Bruton's tyrosine kinase (BTK), a mediator of B cell receptor signaling, has been effectively exploited as a therapeutic target in Chronic lymphocytic leukemia (CLL). Ibrutinib is a BTK inhibitor that has shown excellent efficacy in treatment-naïve, relapsed/refractory, and high-risk CLL. However, concerns regarding ibrutinib resistance have emerged and characterizing the underlying mechanisms may aid in identifying patients with increased risk of resistance/disease progression. Moreover, understanding the interplay between determinants of ibrutinib resistance may reveal alternate therapeutic targets. Using a computational biology modeling (CBM) simulation-based method, we reverse-engineered the salient molecular architecture of a CLL patient progressing on ibrutinib to identify potential drug combinations to overcome drug resistance and induce remission.
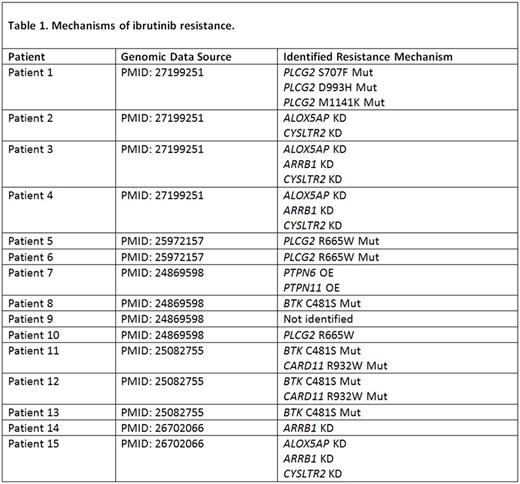
Methods: For validation of the computational biological modeling (CBM) software (Cellworks group), we selected a cohort of 15 ibrutinib resistant CLL patients from previously published studies (Table 1). Genomic data derived from whole exome sequencing (WES), targeted next-generation sequencing (NGS), copy number variation (CNV), and/or karyotype data were used in this study. All available genomic data for each profile was entered into the CBM, which generates a disease-specific protein network map using PubMed and other online resources. Disease-biomarkers that are unique to each patient were identified within the protein network maps by using CBM technology. Ibrutinib was selected from a digital drug library and simulated on every relapsed CLL patient case. Drug impact was assessed by quantitatively measuring the drug's effect on a cell growth score, which is a composite of cell proliferation, viability, and apoptosis, along with the simulated impact on each disease-specific biomarker score. Each patient-specific map was digitally screened to identify any Ibrutinib resistance mechanism(s).
Results: In our modeled patient set, 4 different resistance mechanisms were identified across 14/15 ibrutinib- refractory CLL cases. 1) BTK C481S mutation (n=4) impacts ibrutinib binding affinity; 2) PLCG2 mutations (n=4) lead to activation of PRKCB and AKT1 signaling; 3) CARD11 mutation (n=2) results in constitutive activation of BTK downstream effectors; and 4) decreased G-protein coupled receptor (GPCR) pathway signaling, an upstream activator of BTK (n=5), was found to be a novel resistance mechanism identified by CBM. Knockdown (KD) of ALOX5AP, ARRB1 and CYSLTR2 reduced GPCR pathway signaling, resulting in reduced BTK activity (Table 1). After validating the CBM algorithm, we rendered a digital avatar for a relapsed-refractory CLL patient (Del17p+); progressing on ibrutinib. The patient had rapidly escalating ALC and lymphadenopathy; with the largest node measuring ~16cm. In addition to p53 loss, simulation modeling accounted for BTK C481S mutation (BAF 0.69) expressed in the patients tumor cells. CBM predicted venetoclax would be the most effective therapeutic for the patient due to several upstream signaling pathways converging on Bcl-2. Indeed, addition of venetoclax to the ongoing ibrutinib monotherapy dramatically reduced the patients ALC (from 113 to 3.3 x 109/L) by day 4 post-treatment, accompanied by reduction in lymphadenopathy (left axillary node decreased to ~3cm). Ibrutinib was discontinued within 1 month of starting the combination, with venetoclax monotherapy continued per dosing guidelines thereafter. Notably, the patient maintained remission status on venetoclax monotherapy alone.
Conclusions: Computational modeling was used to identify ibrutinib resistance mechanisms in CLL patients. Using this novel approach, we identified an effective therapy (venetoclax) for a high-risk CLL patient who was progressing on ibrutinib. CBM-based approaches can be used to predict alternative therapeutic regimens for drug-refractory patients, targeting the disease-specific biomarkers unique to each patient. Altogether, our study demonstrates proof-of-principle on the use of computational predictive algorithms to simulate multiple protein-interaction networks operational in a tumor cell to exploit individualized therapeutic vulnerabilities.
Ysebaert: Janssen: Consultancy, Research Funding, Speakers Bureau. Kumar: Cellworks: Employment. Singh: Cellworks Research India Pvt. Ltd: Employment. Ullal: Cellworks Research India: Employment. Bhargav: Cellworks: Employment. Azam: cellworks: Employment. Abbasi: Cellworks Group Inc.: Employment. Vali: Cellworks Group Inc.: Employment. Cogle: Celgene: Other: Membership on Steering Committee for Connect MDS/AML Registry.
Author notes
Asterisk with author names denotes non-ASH members.