Key Points
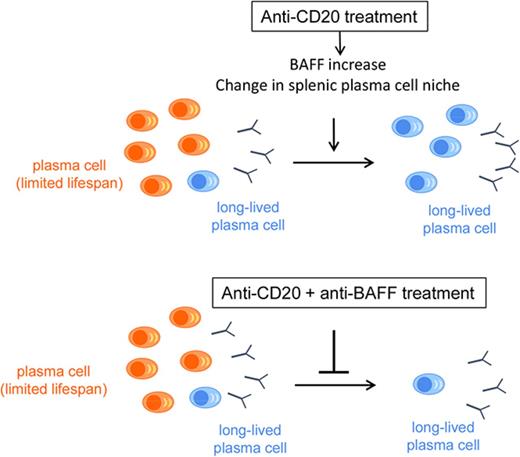
Modification of the splenic microenvironment induced by B-cell depletion creates a dependence of PCs on BAFF and CD4+ T cells.
Combining anti-CD20 and anti-BAFF reduces the number of splenic PCs, opening therapeutic perspectives for antibody-mediated cytopenia.
Abstract
Previous data have suggested that B-cell–depletion therapy may induce the settlement of autoreactive long-lived plasma cells (LLPCs) in the spleen of patients with autoimmune cytopenia. To investigate this process, we used the AID-CreERT2-EYFP mouse model to follow plasma cells (PCs) engaged in an immune response. Multiplex polymerase chain reaction at the single-cell level revealed that only a small fraction of splenic PCs had a long-lived signature, whereas PCs present after anti-CD20 antibody treatment appeared more mature, similar to bone marrow PCs. This observation suggested that, in addition to a process of selection, a maturation induced on B-cell depletion drove PCs toward a long-lived program. We showed that B-cell activating factor (BAFF) and CD4+ T cells play a major role in the PC survival niche, because combining anti-CD20 with anti-BAFF or anti-CD4 antibody greatly reduce the number of splenic PCs. Similar results were obtained in the lupus-prone NZB/W model. These different contributions of soluble and cellular components of the PC niche in the spleen demonstrate that the LLPC expression profile is not cell intrinsic but largely depends on signals provided by the splenic microenvironment, implying that interfering with these components at the time of B-cell depletion might improve the response rate in autoimmune cytopenia.
Introduction
Plasma cells (PCs) are terminally differentiated B cells producing antibodies that provide immediate and long-term protection against pathogens. Circulating high-affinity antibodies are sustained for decades by long-lived plasma cells (LLPCs) that mainly reside in the bone marrow.1-5 PC longevity depends on a supportive environment that forms a specific survival niche, with the number of LLPCs likely being limited by the available space in the niche. In autoimmune contexts, the spleen and inflamed tissues can provide an additional survival niche for autoreactive LLPCs, as shown in the lupus-prone NZB/NZW mouse.6 LLPCs have also been described in the spleens of normal mice in the context of B-cell depletion induced by irradiation or with anti-CD20 therapy.3-5 However, little is known about the cellular environment and signals required for the establishment of such LLPCs in the spleen.
Immune thrombocytopenia (ITP) and warm autoimmune hemolytic anemia (wAIHA) are mediated by pathogenic autoantibodies, mainly produced in the spleen, targeting platelets in ITP and red blood cells in wAIHA. As for a wide range of autoimmune disorders, the use of rituximab, a chimeric anti-CD20 monoclonal antibody, has become a mainstay in the therapy of these 2 diseases, albeit with variable outcomes given that ∼40% of patients with ITP and 30% with wAIHA fail to respond to rituximab, with splenectomy performed as a second-line treatment.7
Our previous data suggested that, in the context of ITP and wAIHA, the use of rituximab paradoxically induced the settlement of splenic LLPCs, some of them being autoreactive, with LLPCs surprisingly less frequent in the spleens of untreated patients.8,9 The presence of this LLPC population could explain the rituximab treatment failure for most of the patients and their subsequent response to splenectomy. The supernatant of spleen cell cultures from ITP and wAIHA patients who received rituximab showed an increased level of B-cell-activating factor (BAFF),8,9 a member of the tumor necrosis factor family with major B-cell survival function, which concurs with previous observations of increased serum levels of BAFF under conditions of B-cell depletion.10,11 Hence, B-cell depletion induced by rituximab may promote new environmental conditions suitable for the maturation and survival of autoimmune LLPCs in the spleen.
In this study, we took advantage of the knockin transgenic mouse model AID-Cre-ERT2xRosa26-loxP-EYFP, which allows the irreversible expression of the fluorescent EYFP marker in B cells engaged in an immune response on a tamoxifen regimen to follow PCs at different times after immunization.12 We characterized the PC maturation stage at the single-cell level on B-cell depletion in the spleen and in bone marrow. We provide a mechanistic view of the process that leads to the differentiation into LLPCs in the spleen, identify major cytokines and cellular components involved in this process, and demonstrate that treatments targeting the PC niche, combined with B-cell depletion, can have a major impact on PC survival in this organ.
Methods
Mice and treatments
AID-Cre-ERT2xRosa26-loxP-EYFP mice (AID-Cre-EYFP throughout this paper) have been previously described.12 Female NZB/NZW mice were purchased from Envigo. All mice were bred and maintained under specific-pathogen–free conditions. Animal experiments and protocols were approved by the Paris Descartes Ethical Committee and the French Ministery of Research. Treatment protocols are summarized in supplemental Figures 2 and 14 and supplemental Methods (available on the Blood Web site).
Flow cytometry and cell sorting
Cells were suspended in Hanks balanced salt solution supplemented with 10% fetal bovine serum and labeled with primary and secondary antibodies for 15 minutes at 4°C (supplemental Table 2). Cell viability was determined with Sytox Live Dead (Life Technologies). Intracellular staining was performed after cell fixation and permeabilization with BD Cytofix/Cytoperm solution. Stained cells were analyzed with a FACS Fortessa apparatus (Becton Dickinson) and DIVA software. Cell sorting involved a FACS Aria cell sorter (BD). EYFP+ PCs were first enriched by a first sorting and then purified by a second one. Cell purity after the second sorting was >99%.
Gene expression profiling
Mouse splenic plasmablasts (PBs)/PCs (EYFP+B220−CD138+) were sorted 6 days after a third immunization with sheep red blood cells (SRBCs) (boosted mice), and mature PCs (EYFP+B220−) were sorted 3 months after the second immunization (control mice). For gene expression profiling, RNA was isolated from sort-purified PCs from 3 boosted mice and 4 control mice (see supplemental Methods).
Single-cell gene expression analysis
Single-cell gene expression analysis was performed on EYFP+B220− PCs and PBs from AID-Cre-EYFP mice. We sorted splenic PBs 3 days after a boost with SRBCs from 2 mice, splenic PCs (Spl-PCs) and bone-marrow PCs (BM-PCs) 3 months after the second immunization from 2 control mice, and Spl-PCs from 4 anti-CD20–treated mice (anti-CD20 PCs). PCs from NZB/W mice were also analyzed at the single-cell level. After dump staining (CD3/CD4/Ly6G/CD11b), we sorted 2 Spl-PC populations (CD138highB220low and CD138highB220−), BM-PCs (CD138highB220−) from 2 27-week-old untreated mice, and Spl-PCs (CD138highB220−) from 4 anti-CD20–treated mice. Single-cell gene expression analysis was performed as previously described (supplemental Methods). Assays and complementary DNA (cDNA) mixes were then transferred to a 48.48 Dynamic Array primed chip, and real-time PCR (RT-PCR) was run according to the Fluidigm protocol. We used 100-cell and no-cell wells as positive and negative controls, respectively. Data were analyzed by using Fluidigm Real-Time PCR analysis software. CD3 gene was used as negative control. Cells expressing B2m and expressing the PC master gene Prdm1 were considered for further analysis. Gene expression was considered positive when detectable under 25 cycles.
Confocal microscopy of spleens
Confocal microscopy of spleens was performed as previously described (supplemental Methods). Images were acquired with LSM 700 (Zeiss) or SP5 (Leica) microscope. Images were taken with a 40× objective and were analyzed and processed by using ImageJ software version 1.46.
Cytokine and immunoglobulin ELISA
Culture supernatants of 4 × 106 spleen cells plated in 24-well plates were collected on day 3. Quantitative determination of BAFF level in sera and of BAFF and APRIL in supernatants of splenocytes involved the use of a mouse BAFF enzyme-linked immunosorbent assay (ELISA) kit (Quantikine, R&D systems) or a mouse APRIL ELISA kit (Lsbio). Immunoglobulin G1 (IgG1), IgA, and IgM levels were determined in sera before and after treatments using Multiscreen HTS 96-well plates (Millipore) coated with 10 µg/mL goat anti-mouse Ig, and revealed with horseradish peroxidase–conjugated anti-mouse Ig antibodies (supplemental Table 2).
Quantification of gene expression by RT-PCR
Total RNA was extracted with the RNeasy Microkit (Qiagen). RNA was reverse-transcribed with the Reverse Transcriptase Kit (Agilent). For EYFP+ cell samples, cDNA was preamplified using Single Primer Isothermal Amplification (Nugen). Gene expression was measured by quantitative RT-PCR. Gene expression was normalized to B2m expression. Taqman primers are listed in supplemental Table 3.
In vitro assays
After sorting, splenic EYFP+B220− PCs were cocultured in 96-well plates with CD4+ T cells from immunized mice, not treated with anti-CD20 or 5 days after the second anti-CD20 injection (CD4+ T-cell–to-PC ratio, 5:1). Cells were cultured in complete RPMI (Gibco) supplemented with 10% fetal bovine serum. Cells were collected at day 5, and PC survival was estimated by fluorescence-activated cell sorting as the number of Sytox−Annexin− cells (BD Biosciences kit).
Data availability
The microarray data are available at https://www.ebi.ac.uk/arrayexpress/ under accession number E-MTAB-6449.
Results
PC maturation after immune responses in the spleens of AID-Cre-EYFP mice
Cell-intrinsic labeling models are powerful tools to analyze PCs, which are rare populations in vivo with no universal surface marker. To this end, we used the AID-Cre-ERT2xROSA26-loxP-EYFP mouse line (hereafter, AID-Cre-EYFP) to follow the fate of PCs generated during an immune response. In this mouse, tamoxifen ingestion coupled with immunization induces the timely labeling of B cells engaged in a germinal center (GC) response, thus allowing the follow-up of persistent memory and PC populations through their acquired EYFP expression (supplemental Figure 1).12,13 A prime-boost immunization was performed with 2 SRBC injections 1 month apart and 3 tamoxifen labeling episodes. A third immunization was also performed to study newly generated PCs a few days after this new challenge (supplemental Figure 2A).
Spl-PCs, identified as EYFP+B220− (Figure 1A), expressed TACI and heterogeneous levels of CD138 and EpCAM (supplemental Figure 3). Single-cell analysis (see below) confirmed that most EYFP+B220− cells are authentic PCs by their expression of Prdm1 and/or Xbp1. To analyze transcriptional changes between newly generated PBs/PCs and mature PCs, we sorted splenic EYFP+B220− PB/PCs 6 days after a third immunization with SRBCs, and splenic EYFP+B220− PCs 3 months after the second immunization (Figure 1A). PC numbers showed a 30-fold expansion between the day-6 boost and the 3-month samples, which reflects the strong proliferative burst taking place in the recall response (Figure 1B).
Gene expression profiling of newly generated and mature splenic EYFP+PCs from mice. Splenic newly generated EYFP+B220− PCs were sorted 6 days after a third immunization with SRBCs (day 6 [D6]/boost), and splenic mature EYFP+B220− PCs 3 months after the second immunization (M3/controls) (see protocol in supplemental Figure 2A). About 10 000 EYFP+ PCs per mouse were analyzed (D6, n = 3; M3, n = 4). (A) Sorting strategy of mature splenic EYFP+ B220− PCs by use of a 2-step procedure. (B) Total number of EYFP+B220− PCs in controls (M3) or boosted (D6) mice. (C) Heatmap of genes selected from the supervised microarray comparison of newly generated (D6) vs mature PCs (M3) in the mouse spleen (223 genes with fold change in expression >4 or <0.25, P < .05). The heatmap is based on normalized log2 intensities that were probe-wise mean-centered across the 2 compared conditions. Columns represent individual samples, and rows represent specific gene probes, with upregulated genes in red and downregulated genes in blue. Genes selected for the single-cell analysis are highlighted. (D) Functional analysis of the 174 upregulated genes in mature vs newly generated PCs (more than fourfold upregulated; P < .05). The number of genes in each functional category is shown.
Gene expression profiling of newly generated and mature splenic EYFP+PCs from mice. Splenic newly generated EYFP+B220− PCs were sorted 6 days after a third immunization with SRBCs (day 6 [D6]/boost), and splenic mature EYFP+B220− PCs 3 months after the second immunization (M3/controls) (see protocol in supplemental Figure 2A). About 10 000 EYFP+ PCs per mouse were analyzed (D6, n = 3; M3, n = 4). (A) Sorting strategy of mature splenic EYFP+ B220− PCs by use of a 2-step procedure. (B) Total number of EYFP+B220− PCs in controls (M3) or boosted (D6) mice. (C) Heatmap of genes selected from the supervised microarray comparison of newly generated (D6) vs mature PCs (M3) in the mouse spleen (223 genes with fold change in expression >4 or <0.25, P < .05). The heatmap is based on normalized log2 intensities that were probe-wise mean-centered across the 2 compared conditions. Columns represent individual samples, and rows represent specific gene probes, with upregulated genes in red and downregulated genes in blue. Genes selected for the single-cell analysis are highlighted. (D) Functional analysis of the 174 upregulated genes in mature vs newly generated PCs (more than fourfold upregulated; P < .05). The number of genes in each functional category is shown.
By supervised microarray analysis comparing mature Spl-PCs and newly generated PBs/PCs, we identified 223 differentially expressed genes (expression fold-change >4 or <0.25, P < .05), among which 174 genes were upregulated in mature PCs as compared with PBs/PCs and 49 were downregulated (Figure 1C; supplemental Table 1). We found no cell cycle gene, apart from Chek1, upregulated in newly generated PBs/PCs, indicating that the differentiation of memory B cells into antibody-secreting cells (ASCs) in a recall response involves a rapid exit of the cell cycle.
Upregulated genes in newly generated PBs/PCs included Cxcr3, involved in cell migration, Bcl2l11 (Bim), a proapoptotic factor, and several activators of signal transduction (Bank1, Smad3, and Plaa). Upregulated genes in the 3-month-old PC population included antiapoptotic factors (Tnfaip3 and Bcl2), transcription factors (Nfkb2, Runx2, Irf8, Tox2, and Klf2), regulators of signal transduction (Btla), membrane receptors (Tnfrsf17, coding for Bcma, Ctla4, and IcosL), and numerous genes involved in ion transport (Tesc, Clic5, and S100a6), intracellular trafficking (Rab27a and several Gimap genes), and metabolic pathways (Mt2, Anxa2, Cox15, and Atp1b1) (Figure 1C-D). The 2 populations did not differ in the expression of transcription factors involved in PC identity, such as Xbp1, Irf4, jun, or Ell2, whereas Cited2, recently identified by Shi et al, showed a twofold increase in mature PCs (supplemental Figure 4).14 Prdm1 (encoding Blimp-1) showed a threefold expression difference, reflecting the maturation occurring between these 2 subsets that constitutes the basis of PC discrimination in the Blimp-EGFP reporter mouse.14 Hence, recently formed, nondividing PCs further undergo important transcriptional changes during their maturation.
Follow-up of PCs during B-cell depletion in AID-Cre-EYFP mice
Mice were treated or not with anti-CD20 1 month after the second immunization (supplemental Figure 2B). Analysis 4 hours after 1 injection of anti-CD20 showed that 76% of B cells and 11% of PCs were strongly stained by this antibody (supplemental Figure 5). B-cell depletion in the spleen was maximal at 50 days after the first anti-CD20 injection (ie, 3 months after the last immunization), with ∼2% of splenic residual B cells (Figure 2A-B). At that time, B cells were scarcely detectable in peripheral blood. PCs, identified as EYFP+B220−, represented the major EYFP subset that resisted B-cell depletion. The number of EYFP+B220− PCs was slightly but not significantly decreased in anti-CD20–treated mice as compared with controls (Figure 2C), which could correlate with the PC fraction marked by the anti-CD20 antibody. Among all EYFP+ PCs, only IgG1-positive cells showed a significant decrease (2.5-fold) after anti-CD20 antibody treatment (Figure 2D), whereas IgM and IgA were moderately changed, thus accounting for the modest alteration in total PC numbers.
Impact of B-cell depletion in the spleen of AID-Cre-EYFP mice. AID-Cre-EYFP mice were immunized by 2 SRBCs injections 1 month apart, together with a tamoxifen regimen, and were injected with (n = 14) or without (n = 12) anti-CD20 antibody (3 × 250 µg, IV) (see supplemental Figure 2B). Analysis originates from ≥2 independent tamoxifen-labeling experiments. (A) Kinetics of B-cell depletion in mouse blood (D0 represents the first day of anti-CD20 antibody injection). (B) Comparison of EYFP+B220+ B-cell numbers in spleens of control mice (3 months after immunization) and anti-CD20–treated mice at the nadir of B-cell depletion (day 50 after first anti-CD20 injection). (C) Comparison of EYFP+B220− PC numbers per spleen in controls and anti-CD20–treated mice at day 50 after the first anti-CD20 antibody injection. (D) Comparison of IgG1+, IgA+, and IgM+ splenic EYFP+B220− PC numbers in controls (n = 4) and anti-CD20–treated (n = 4) mice at day 50 after the first anti-CD20 injection, determined by intracellular staining. Anti-CD20–treated and control mice showed a 2.5-fold difference in the number of IgG1+ cells (P < .01). Significant differences were estimated by the Mann-Whitney U test (****P < .001). Mean values are indicated.
Impact of B-cell depletion in the spleen of AID-Cre-EYFP mice. AID-Cre-EYFP mice were immunized by 2 SRBCs injections 1 month apart, together with a tamoxifen regimen, and were injected with (n = 14) or without (n = 12) anti-CD20 antibody (3 × 250 µg, IV) (see supplemental Figure 2B). Analysis originates from ≥2 independent tamoxifen-labeling experiments. (A) Kinetics of B-cell depletion in mouse blood (D0 represents the first day of anti-CD20 antibody injection). (B) Comparison of EYFP+B220+ B-cell numbers in spleens of control mice (3 months after immunization) and anti-CD20–treated mice at the nadir of B-cell depletion (day 50 after first anti-CD20 injection). (C) Comparison of EYFP+B220− PC numbers per spleen in controls and anti-CD20–treated mice at day 50 after the first anti-CD20 antibody injection. (D) Comparison of IgG1+, IgA+, and IgM+ splenic EYFP+B220− PC numbers in controls (n = 4) and anti-CD20–treated (n = 4) mice at day 50 after the first anti-CD20 injection, determined by intracellular staining. Anti-CD20–treated and control mice showed a 2.5-fold difference in the number of IgG1+ cells (P < .01). Significant differences were estimated by the Mann-Whitney U test (****P < .001). Mean values are indicated.
Single-cell gene expression analysis of PCs from anti-CD20–treated mice reveals a long-lived gene expression profile
From our transcriptomic analysis, we defined a set of genes that could allow us to distinguish, at the single-cell level, the process of PC maturation. For LLPC genes, we selected Tnfaip3, Bcl2, Runx2, Nfkb2, Tox2, Mt2, Tesc, and Tnfrsf17, which showed increased expression in the 3-month PC samples. We included Tmem176b, highlighted by Shi et al as a PC maturation marker, which showed a 3.6 difference in our microarray analysis.14 We also selected Klf6, which had a relevant value in our previous analysis of human PC maturation.9 For plasmablast genes, we selected Chek1, Bcl2l11, and Cxcr3, the 3 most relevant ones from our analysis of PBs/PCs at day 6, and included 5 other genes linked with cell proliferation: Mki67, Bub1, Aurka, Ccnd2, and Rrm2b. Expression of Mki67, Bcl2, and Nfkb2 was evaluated by quantitative RT-PCR on splenic EYFP+ PBs/PCs, mature PCs, and PCs from anti-CD20–treated mice, which showed a significant decrease (Mki67) or a small variation (Bcl2 and Nfkb2) between the last 2 populations (supplemental Figure 6). We also observed that Tnfaip3 expression was increased at the protein level after anti-CD20 treatment in Spl-PCs (supplemental Figure 7).
We sorted newly generated splenic PBs at an earlier stage (ie, 3 days after a boost with SRBCs [“boost”]); Spl-PCs and BM-PCs were sorted 3 months after the second immunization and Spl-PCs from anti-CD20–treated mice at the nadir of B-cell depletion (day 50) (anti-CD20 PCs). Most newly generated splenic PBs expressed Ki67 protein (supplemental Figure 8). Splenic EYFP+ PCs were sorted by using the EYFP+B220− criteria in all cases. Single-cell gene expression profiling was performed by multiplex RT-PCR, and the results were visualized by heatmap and analyzed by principal component analysis (Figure 3A-B). A total of 3.5% of all cells analyzed were negative for both Prdm1 and Xbp1, and 12.7% were negative for Prdm1 alone. These cells were excluded from further analyses.
Maturation of splenic EYFP+PCs on B-cell depletion. (A) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on PBs, Spl-PCs, anti-CD20 PCs, and BM-PCs using 18 genes (PB genes: Mki67, Chek1, Bub1, Aurka, Rrm2b, Ccnd2, Bcl2l11, and Cxcr3; LLPC genes: Tesc, Klf6, Tnfaip3, Bcl2, Runx2, Tnfrsf17, Nfkb2, Tox2, Tmem176b, and Mt2) discriminating PBs from LLPCs. Analysis was restricted to cells expressing B2m and the Prdm1 PC master gene. Columns represent individual gene probes, and rows represent single cells, with color relative to the cycle threshold expression value (scale bar at bottom). (B) Principal component analysis of 441 individual cells analyzed for the expression of 18 diagnostic genes. Each dot represents a single cell: gray, splenic ASCs generated 3 days after a third immunization (boost); blue, PCs 3 months after immunization (Spl-PCs); orange, PCs at the nadir of B-cell depletion (anti-CD20 PCs); green, BM-PCs 3 months after the second immunization (same animals as Spl-PCs). Multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array involved 140 cells per group (C) Percentage of individual cells expressing n diagnostic PC genes in each population (boost [gray], Spl-PCs [blue], anti-CD20 PCs [orange], BM-PCs [green]). (D) Cumulative percentage of cells expressing n or more PC genes in each group.
Maturation of splenic EYFP+PCs on B-cell depletion. (A) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on PBs, Spl-PCs, anti-CD20 PCs, and BM-PCs using 18 genes (PB genes: Mki67, Chek1, Bub1, Aurka, Rrm2b, Ccnd2, Bcl2l11, and Cxcr3; LLPC genes: Tesc, Klf6, Tnfaip3, Bcl2, Runx2, Tnfrsf17, Nfkb2, Tox2, Tmem176b, and Mt2) discriminating PBs from LLPCs. Analysis was restricted to cells expressing B2m and the Prdm1 PC master gene. Columns represent individual gene probes, and rows represent single cells, with color relative to the cycle threshold expression value (scale bar at bottom). (B) Principal component analysis of 441 individual cells analyzed for the expression of 18 diagnostic genes. Each dot represents a single cell: gray, splenic ASCs generated 3 days after a third immunization (boost); blue, PCs 3 months after immunization (Spl-PCs); orange, PCs at the nadir of B-cell depletion (anti-CD20 PCs); green, BM-PCs 3 months after the second immunization (same animals as Spl-PCs). Multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array involved 140 cells per group (C) Percentage of individual cells expressing n diagnostic PC genes in each population (boost [gray], Spl-PCs [blue], anti-CD20 PCs [orange], BM-PCs [green]). (D) Cumulative percentage of cells expressing n or more PC genes in each group.
PBs, collected 3 days after the boost, expressed multiple genes linked with cell division, which confirmed the rapid exit of the cell cycle observed at day 6. BM-PCs sorted 3 months after the second immunization expressed multiple LLPC genes, with ≥4 genes expressed by two-thirds of cells (Figure 3C-D). In contrast, a major fraction of mature PC (Spl-PCs), although not harboring a proliferative profile, expressed a much lower number of PC-specific genes per cell (14% of cells expressing ≥4 genes). PCs isolated after anti-CD20 treatment differed clearly from Spl-PCs in the number of PC genes expressed per cell unit (56% of cells expressing ≥4 genes), similarly to long-lived BM-PCs (64%) (Figure 3C-D). The proximity between the different PC subsets was further validated by the Euclidian distance of their gene expression profile (supplemental Figure 9). Thus, anti-CD20 PCs, which persisted over time since their initial AID-mediated labeling, appear to be composed of a mature population expressing a more robust long-lived program compared with PCs isolated from untreated animals at a similar distance from their EYFP labeling.
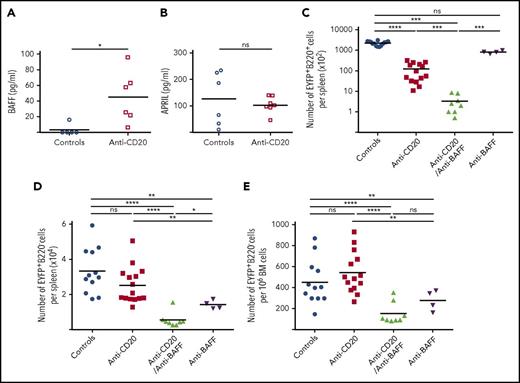
BAFF plays a major role in promoting the emergence of splenic LLPCs
The BAFF cytokine is a major factor in B-cell homeostasis, including at different stages of their activation.15,16 BAFF serum levels increased rapidly after the first anti-CD20 injection (supplemental Figure 10A). We also observed increased BAFF secretion in the culture supernatant of splenocytes isolated from B-cell–depleted animals (Figure 4A), likely due to lower consumption by B cells and increased availability,9-11,17 whereas APRIL secretion did not significantly change (Figure 4B). We therefore chose to target BAFF concomitantly with B-cell depletion and treated immunized mice with anti-BAFF antibody associated or not with anti-CD20 (supplemental Figure 2B). The combined anti-BAFF/anti-CD20 treatment efficiently prevented the increased BAFF serum level (supplemental Figure 10B) and more efficiently depleted splenic B220+EYFP+ B cells than anti-CD20 alone (Figure 4C). Moreover, the combined treatment greatly decreased the number of splenic EYFP+B220− PCs compared with controls (or with anti-CD20), an effect that was more pronounced than with anti-BAFF alone (Figure 4D). The number of EYFP+ BM-PCs decreased threefold as compared with controls, a reduction that more strongly affected IgM PCs (Figure 4E; supplemental Figure 10C). Accordingly, the level of IgM significantly decreased in the sera up to day 70, whereas IgG1 and IgA levels remained less affected (supplemental Figure 10D-I). However, the reduction in EYFP+ BM-PCs observed with the combined treatment was not significantly more pronounced than with BAFF alone. These results demonstrate that blocking BAFF function can interfere with the overall survival of LLPCs in B-cell depleted spleens without a major impact on LLPCs with a switched isotype in bone marrow.
BAFF is a key survival factor for Spl-PCs on B-cell depletion in AID-Cre-EYFP mice. (A) Quantification of BAFF concentration in culture supernatants of 4 × 106 total spleen cells from control (n = 5) and anti-CD20–treated mice (n = 6) after 3 days (ELISA). (B) Quantification of APRIL concentration in culture supernatants of 4 × 106 total spleen cells from control (n = 6) and anti-CD20–treated mice (n = 7) after 3 days (ELISA). (C-E) Number of residual EYFP+ cells observed at the nadir of B-cell depletion (or at equivalent time points) in controls (n = 12), anti-CD20– (n = 14), anti-CD20/anti-BAFF– (n = 8), and anti-BAFF–treated (n = 4) mice: EYFP+ B220+ splenic B cells (C), EYFP+ B220− Spl-PCs (D) and EYFP+B220− BM-PCs (E) (see protocol in supplemental Figure 2B). Analysis originates from ≥2 independent tamoxifen-labeling experiments, except for the anti-BAFF group. Significant differences are estimated by Mann-Whitney U test or 1-way ANOVA, Dunn test for multiple comparisons (*P < .05; **P < .01; ***P < .001; ****P < .0001). Mean values are indicated.
BAFF is a key survival factor for Spl-PCs on B-cell depletion in AID-Cre-EYFP mice. (A) Quantification of BAFF concentration in culture supernatants of 4 × 106 total spleen cells from control (n = 5) and anti-CD20–treated mice (n = 6) after 3 days (ELISA). (B) Quantification of APRIL concentration in culture supernatants of 4 × 106 total spleen cells from control (n = 6) and anti-CD20–treated mice (n = 7) after 3 days (ELISA). (C-E) Number of residual EYFP+ cells observed at the nadir of B-cell depletion (or at equivalent time points) in controls (n = 12), anti-CD20– (n = 14), anti-CD20/anti-BAFF– (n = 8), and anti-BAFF–treated (n = 4) mice: EYFP+ B220+ splenic B cells (C), EYFP+ B220− Spl-PCs (D) and EYFP+B220− BM-PCs (E) (see protocol in supplemental Figure 2B). Analysis originates from ≥2 independent tamoxifen-labeling experiments, except for the anti-BAFF group. Significant differences are estimated by Mann-Whitney U test or 1-way ANOVA, Dunn test for multiple comparisons (*P < .05; **P < .01; ***P < .001; ****P < .0001). Mean values are indicated.
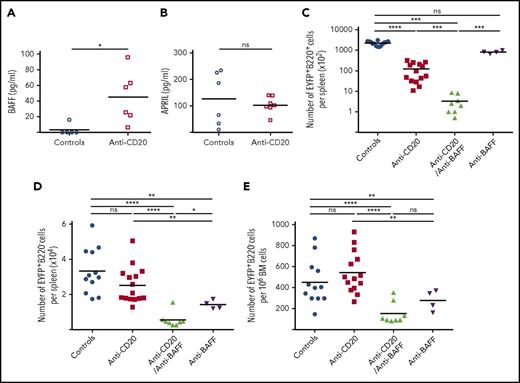
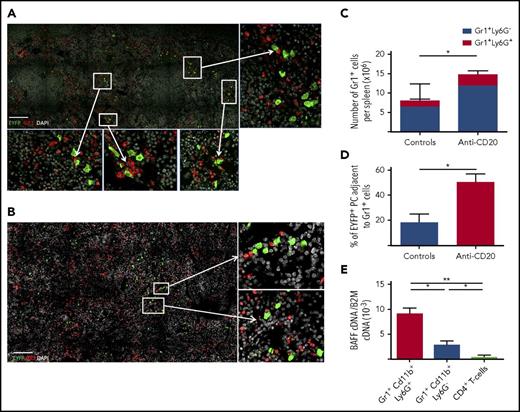
BAFF-producing neutrophils are components of the Spl-PC niche
EYFP+ PCs can be easily discriminated from memory B cells by confocal microscopy through their strong EYFP cytoplasmic staining (supplemental figure 11). Gr1+ cells (including neutrophils, monocytes, and dendritic cells) were found in close proximity to Spl-PCs in the red pulp (Figure 5A-B). The number of splenic Gr1+ cells increased with B-cell depletion, possibly because of increased space availability for this population with a fast turnover, and the number of Gr1+ cells adjacent with PCs increased consequently (Figure 5C-D). Further analysis confirmed that most Gr1+ cells observed by confocal microscopy were Ly6G+ neutrophils, because Ly6G+ cells are Gr1high (supplemental Figure 12B). No interaction was observed with F4/80+ macrophages or FcεRI+ basophils (data not shown). Gr1+CD11b+Ly6G+ neutrophils expressed higher BAFF levels compared with Gr1+Cd11b+Ly6G− monocytes and dendritic cells or CD4+ T cells (Figure 5E). Therefore, we aimed to deplete neutrophils by repeated injections of an anti-Ly6G antibody over 7 weeks (supplemental Figure 2C). However, prolonged treatment with anti-Ly6G antibody did not affect BAFF serum level, but induced the expansion of a Gr1+CD11b+Ly6G− population that expressed BAFF at a level similar to that in Ly6G+ neutrophils (supplemental Figure 13A-B). The Ly6G-depleting treatment may have forced the emergence of a neutrophil population that reduced Ly6G surface expression while maintaining its BAFF secretion capacity, thus rendering the targeting of the neutrophil component of the PC niche inoperative.
Interaction of Spl-PCs with BAFF-producing neutrophils. (A) Interactions of EYFP+ PCs (green) and Gr1+ cells (red) in the spleen of a representative anti-CD20–treated mouse (day 50 after the first anti-CD20 injection) observed by 3-color confocal microscopy with an LSM 700 (Zeiss) and 40× objective. Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bar, 150 µm. PCs can be easily discriminated from memory B cells through their EYFP intensity (see supplemental Figure 11 and supplemental Methods). (B) Same analysis for control mice 3 months after 2 SRBC immunizations. Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bar, 150 µm. (C) Number of Gr1+Ly6G+ and Gr1+Ly6G− cells in spleens from controls 3 months after 2 SRBC immunizations and anti-CD20–treated mice 50 days after the first anti-CD20 injection. (D) Quantification of EYFP+ PC-Gr1+ cell interactions in the spleen of control and B-cell–depleted mice. Spleens from 3 mice per group were analyzed corresponding to about 600 EYFP+ PCs in the control group and 300 EYFP+ PCs in the anti-CD20–treated group. Interactions between EYFP+ PCs and Gr1+ cells were calculated by using ImageJ software. (E) Neutrophils (Gr1+Cd11b+Ly6G+ cells) were the predominant source of BAFF production in the spleen compared with other Gr1+ cells and CD4+ T cells (P < .05). CD4+ T cells, Gr1+Cd11b+Ly6G− cells and Gr1+Cd11b+Ly6G+ cells were collected from the spleen of 3 immunized mice. About 10 000 cells per population for each mouse were sorted. BAFF cDNA expression was quantified by RT-PCR and normalized to B2m expression (2−ΔCt). Significant differences are estimated by Mann-Whitney U test or 1-way analysis of variance, Dunn test for multiple comparisons (*P < .05; **P < .01). Mean values are indicated.
Interaction of Spl-PCs with BAFF-producing neutrophils. (A) Interactions of EYFP+ PCs (green) and Gr1+ cells (red) in the spleen of a representative anti-CD20–treated mouse (day 50 after the first anti-CD20 injection) observed by 3-color confocal microscopy with an LSM 700 (Zeiss) and 40× objective. Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bar, 150 µm. PCs can be easily discriminated from memory B cells through their EYFP intensity (see supplemental Figure 11 and supplemental Methods). (B) Same analysis for control mice 3 months after 2 SRBC immunizations. Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bar, 150 µm. (C) Number of Gr1+Ly6G+ and Gr1+Ly6G− cells in spleens from controls 3 months after 2 SRBC immunizations and anti-CD20–treated mice 50 days after the first anti-CD20 injection. (D) Quantification of EYFP+ PC-Gr1+ cell interactions in the spleen of control and B-cell–depleted mice. Spleens from 3 mice per group were analyzed corresponding to about 600 EYFP+ PCs in the control group and 300 EYFP+ PCs in the anti-CD20–treated group. Interactions between EYFP+ PCs and Gr1+ cells were calculated by using ImageJ software. (E) Neutrophils (Gr1+Cd11b+Ly6G+ cells) were the predominant source of BAFF production in the spleen compared with other Gr1+ cells and CD4+ T cells (P < .05). CD4+ T cells, Gr1+Cd11b+Ly6G− cells and Gr1+Cd11b+Ly6G+ cells were collected from the spleen of 3 immunized mice. About 10 000 cells per population for each mouse were sorted. BAFF cDNA expression was quantified by RT-PCR and normalized to B2m expression (2−ΔCt). Significant differences are estimated by Mann-Whitney U test or 1-way analysis of variance, Dunn test for multiple comparisons (*P < .05; **P < .01). Mean values are indicated.
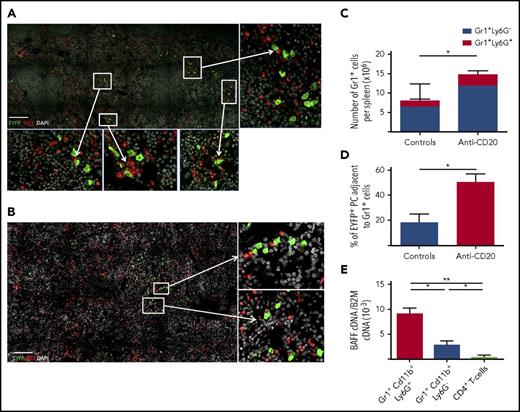
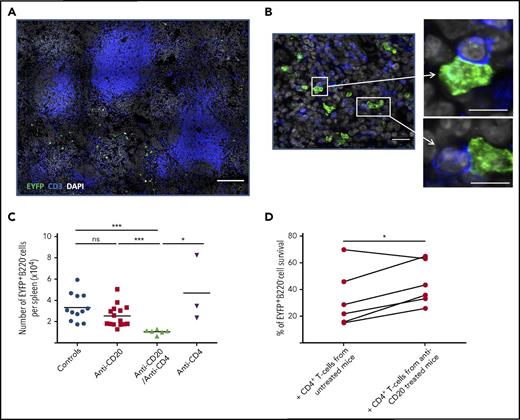
CD4+ T cells contribute to the survival niche of splenic LLPCs in the context of B-cell depletion
By confocal microscopy, we also observed that 20% to 30% of Spl-PCs were in close contact with CD3+ T cells (mostly CD4+, data not shown) in both treated or untreated mice (Figure 6A-B). The number of PC-CD3+ T-cell interactions did not significantly increase with B-cell depletion (median: 20.5% in control group vs 30% in anti-CD20 mice). We used CD4+ T-cell depletion, with or without anti-CD20 treatment, to assess the contribution of CD4+ T cells to PC survival (supplemental Figure 2C). Combining CD4+ T-cell depletion with anti-CD20 antibody treatment, but not CD4+ T-cell depletion alone, significantly decreased the number of Spl-PCs (Figure 6C). The differential effect in the context of B-cell depletion could be due to CD4+ T-cell activation processes induced by BAFF or by other splenic factors modified by anti-CD20 antibody treatment. Hence, we cocultured splenic EYFP+B220− PCs with autologous CD4+ T cells previously primed with various concentrations of BAFF and observed no effect on PC survival (data not shown). However, coculture in vitro with CD4+ T cells isolated from anti-CD20–treated mice slightly but significantly increased the survival of splenic EYFP+B220− PCs as compared with cultures with CD4+ T cells from control animals (Figure 6D). These results suggest that CD4+ T cells might be primed to provide additional signals to Spl-PCs in the context of B-cell depletion, which could contribute to the emergence or survival of LLPCs.
CD4+T-cells contribute to the persistence of splenic LLPCs in anti-CD20–treated AID-Cre-EYFP mice. (A-B) Analysis of interactions between EYFP+ PCs (green) and CD3+ T cells (blue) in the spleen on B-cell depletion. We analyzed total spleen sections from 3 mice by confocal microscopy corresponding to ∼400 EYFP+ PCs in controls and 300 EYFP+ PCs in anti-CD20–treated mice. Images were acquired by confocal microscopy with LSM 700 (Zeiss) and 40× objective. A threshold of confocal resolution allows the unambiguous detection of PCs. (A-B) Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bars, 150 µm (A), 18 µm (B, left panel), 10 µm (B, right panel). (C) Number of EYFP+ B220− PCs per spleen in controls, anti-CD20–, anti-CD20/anti-CD4–, and anti-CD4–treated mice (see supplemental Figure 2C for protocol). Analysis is from 2 independent tamoxifen-labeling experiments except for anti-CD4. (D) Survival of splenic EYFP+ PCs cocultured for 5 days with CD4+ T cells from anti-CD20–treated mice or CD4+ T cells from controls. Splenic EYFP+ PCs were cultured after sorting from 5 immunized mice, with a CD4+ T-cell–to-PC ratio of 5:1. CD4+ T cells were collected from untreated or anti-CD20–treated immunized mice (5 days after the second anti-CD20 injection). Assays were performed in duplicate per condition and per mouse. Significant differences are estimated by paired Student t test or 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001). Mean values are indicated.
CD4+T-cells contribute to the persistence of splenic LLPCs in anti-CD20–treated AID-Cre-EYFP mice. (A-B) Analysis of interactions between EYFP+ PCs (green) and CD3+ T cells (blue) in the spleen on B-cell depletion. We analyzed total spleen sections from 3 mice by confocal microscopy corresponding to ∼400 EYFP+ PCs in controls and 300 EYFP+ PCs in anti-CD20–treated mice. Images were acquired by confocal microscopy with LSM 700 (Zeiss) and 40× objective. A threshold of confocal resolution allows the unambiguous detection of PCs. (A-B) Spleen sections were stained with anti-EYFP, anti-CD3 antibodies, and DAPI. Scale bars, 150 µm (A), 18 µm (B, left panel), 10 µm (B, right panel). (C) Number of EYFP+ B220− PCs per spleen in controls, anti-CD20–, anti-CD20/anti-CD4–, and anti-CD4–treated mice (see supplemental Figure 2C for protocol). Analysis is from 2 independent tamoxifen-labeling experiments except for anti-CD4. (D) Survival of splenic EYFP+ PCs cocultured for 5 days with CD4+ T cells from anti-CD20–treated mice or CD4+ T cells from controls. Splenic EYFP+ PCs were cultured after sorting from 5 immunized mice, with a CD4+ T-cell–to-PC ratio of 5:1. CD4+ T cells were collected from untreated or anti-CD20–treated immunized mice (5 days after the second anti-CD20 injection). Assays were performed in duplicate per condition and per mouse. Significant differences are estimated by paired Student t test or 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001). Mean values are indicated.
B-cell depletion induces the emergence of LLPCs in the spleen of NZB/NZW lupus-prone mice, and targeting BAFF reduces this differentiation process
We assessed whether B-cell depletion could also modify the Spl-PC program in lupus-prone NZB/NZW mice. In this model, autoreactive GC B cells start to feed the Spl-PC pool between 6 and 12 weeks, generating both proliferating PBs and a stable non-dividing PC population.6 Mice were treated or not with anti-CD20 antibody starting from age 20 weeks (supplemental Figure 14). Few residual B cells were detectable in spleens at day 50 after the first anti-CD20 injection as compared with untreated mice (supplemental Figure 15A). We used dump staining and B220/CD138 labeling to identify Spl-PCs. Three different CD138-positive subsets were identified according to their B220 expression: B220+CD138high, B220lowCD138high, and B220−CD138high (Figure 7A). The first subset represented B cells, not PCs, because they did not express Blimp1 or Xbp1 (data not shown). Therefore, we sorted the 2 B220low and B220− CD138high PC subsets as well as BM-PCs (B220−CD138high) from 27-week-old untreated mice and Spl-PCs (B220−CD138high, which is the only subset that remained after B-cell depletion) from anti-CD20–treated mice (Figure 7E; supplemental Figure 15B). Multiplex RT-PCR single-cell analysis of these different populations revealed that the 2 PC subsets from untreated mice were very similar in spite of their differential susceptibility to anti-CD20 treatment and contained a heterogeneous mixture of proliferating PBs and PCs (Figure 7B-C). These Spl-PCs (B220low or B220−) differed from anti-CD20 PCs and BM-PCs in that only 2% to 3% expressed at least 4 PC-specific genes per cell as compared with 33% for anti-CD20 PCs and 25% for BM-PCs (Figure 7D). The absolute number of splenic CD138highB220− PCs was reduced twofold, although not significantly, with anti-CD20 treatment, which suggests again that the anti-CD20 antibody may have induced, in addition to the selection of preexisting LLPCs, a maturation of PCs toward the long-lived expression profile. We finally combined B-cell depletion with BAFF blockade and observed a decrease in the number of B220−CD138high Spl-PCs as compared with control or anti-CD20–depleted mice, whereas anti-BAFF alone had no effect (Figure 7E). BAFF blockade or combined anti-BAFF/anti-CD20 treatment had no significant effect on BM-PC number (supplemental Figure 15C). Hence, similar to our observation in AID-Cre-EYFP mice, B-cell–depleting treatment appears to modify the expression pattern of Spl-PCs toward a more mature profile in lupus-prone mice, and combined BAFF and B-cell depletion efficiently interfered with this maturation process and specifically affected PC survival in the splenic microenvironment.
Splenic LLPCs depend on BAFF in the context of B-cell depletion in NZB/W mice. (A) Gating strategy for sorting ASCs from NZB/W mice for single-cell analysis. ASCs from 27-week-old NZB/W mice were sorted after dump-staining with CD3/CD4/Gr1/Cd11b by a 2-step procedure and separated into CD138highB220− and CD138highB220low fractions. ASCs were collected from the spleen (CD138highB220−, CD138highB220low) and bone marrow (CD138highB220−) from 2 untreated and 4 anti-CD20–treated mice at the nadir of B-cell depletion. (B) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on splenic CD138highB220low and CD138highB220− populations using diagnostic genes allowing the discrimination of PBs from LLPCs as in Figure 3A. (C) Percentage of individual cells expressing n PC genes in each population (CD138highB220low [gray], CD138highB220− cells [blue], CD138highB220− anti-CD20 PCs [purple], BM-PCs [green]). (D) Cumulative percentage of individual cells expressing n or more PC genes in each group. (E) Comparison at the nadir of B-cell depletion of CD138highB220− cell numbers in the spleens of mice. Analysis from 2 independent experiments. Significant differences were estimated by 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001; **** P < .0001). Mean values are indicated.
Splenic LLPCs depend on BAFF in the context of B-cell depletion in NZB/W mice. (A) Gating strategy for sorting ASCs from NZB/W mice for single-cell analysis. ASCs from 27-week-old NZB/W mice were sorted after dump-staining with CD3/CD4/Gr1/Cd11b by a 2-step procedure and separated into CD138highB220− and CD138highB220low fractions. ASCs were collected from the spleen (CD138highB220−, CD138highB220low) and bone marrow (CD138highB220−) from 2 untreated and 4 anti-CD20–treated mice at the nadir of B-cell depletion. (B) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on splenic CD138highB220low and CD138highB220− populations using diagnostic genes allowing the discrimination of PBs from LLPCs as in Figure 3A. (C) Percentage of individual cells expressing n PC genes in each population (CD138highB220low [gray], CD138highB220− cells [blue], CD138highB220− anti-CD20 PCs [purple], BM-PCs [green]). (D) Cumulative percentage of individual cells expressing n or more PC genes in each group. (E) Comparison at the nadir of B-cell depletion of CD138highB220− cell numbers in the spleens of mice. Analysis from 2 independent experiments. Significant differences were estimated by 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001; **** P < .0001). Mean values are indicated.
Discussion
We proposed previously that B-cell depletion, via its impact on the splenic microenvironment, could promote the emergence of LLPCs in the spleen of patients with ITP and wAIHA, which may explain the failure of anti-CD20 therapy in some patients. To investigate this process further, we used the knockin AID-Cre-EYFP mouse model to follow PCs at different times after immunization and on B-cell depletion in the spleen and bone marrow. With this model, we could identify PCs without the need for classical surface markers, such as Syndecan-1, whose expression is heterogeneous and which therefore do not represent universal markers. Moreover, the timing of tamoxifen-induced cell labeling allowed us to date the formation of EYFP+ PCs.
The analysis of the transcriptional program of Spl-PCs at 6 days and 3 months after a recall response highlighted an early acquisition of the PC program, because most transcriptional factors, such as Xbp1 or Irf4, involved in ASC identity were already expressed in newly generated PCs, as were key survival factors, such as Mcl1. It also revealed that their maturation program, with increased expression of Blimp-1, mobilizes various effectors, including transcription factors (Nfkb2 and Runx2), anti-apoptotic genes (Bcl2 and Tnfaip3), and several metabolic or cell signaling pathways (S100a6, Mt2, Btla, and Tnfrsf17).
Using a set of diagnostic genes selected from our transcriptomic analysis and from published data,9,14 we performed single-cell gene expression assays of Spl-PCs from anti-CD20–treated mice, which revealed that Spl-PCs acquired on B-cell depletion an expression profile close to that of normal bone-marrow LLPCs. We would have expected that normal PCs, with EYFP labeling similarly acquired from GC responses initiated 3 months before analysis, would represent resident long-lived cells. In fact, in the control PC sample, the single-cell assay revealed a paucity of expressed genes representing a long-lived signature, whereas anti-CD20 and BM-PCs showed a more robust and reinforced long-lived signature at the individual cell level. In the latter, 56% of cells expressed ≥4 long-lived signature genes as compared with 14% in the control PC sample. This fourfold increase in the fraction of Spl-PCs expressing a long-lived program contrasts with the modest reduction in the absolute number of splenic EYFP+ PCs observed after anti-CD20 treatment, a reduction that likely corresponds to the elimination of PCs expressing CD20. These results suggest that, along with the elimination of a minor subset of immature PCs, an additional process of maturation was induced on B-cell depletion that drove PCs toward a long-lived program.
Several studies have documented an increase in circulating BAFF levels after B-cell–depletion therapies.10,17,18 The BAFF cytokine controls multiple steps of B-cell development and homeostasis by both systemic and local production achieved by different cell subsets.16,19,20 As expected from these previous reports, we observed increased BAFF serum levels as well as increased BAFF secretion in splenic cell cultures of anti-CD20–treated mice. APRIL, one of the main prosurvival cytokines for PCs in the bone marrow, but also for PBs in lymph nodes,21 did not show significant variations in splenic cultures after B-cell depletion. In line with this, simultaneous injection of anti-CD20 and anti-BAFF antibodies markedly reduced the number of splenic EYFP+ PCs as compared with anti-CD20 alone (5-fold). Anti-BAFF treatment alone impacted PC numbers to a lesser extent (twofold), indicating that BAFF contributes to the maintenance of PCs in the spleen. But the major effect observed with the combined treatment suggested that B-cell depletion, by modifying the microenvironment of Spl-PCs and their molecular program, may have created a particular dependence toward BAFF.
The NZB/NZW lupus-prone mouse mirrors many aspects of autoimmune diseases, notably an ongoing GC B-cell response and the generation of short-lived ASCs in the spleen. In this autoimmune context, we also observed a process of maturation on B-cell depletion, and the combined injection of anti-CD20/anti-BAFF antibodies efficiently suppressed Spl-PCs without affecting PC numbers in bone marrow. These results provide a mechanistic explanation for recent reports showing that combining anti-CD20 with BAFF receptor blockade in a prenephritis lupus-prone mouse model enhanced B-cell and PC depletion and reduced progression toward nephritis.22
There is growing evidence that splenic neutrophils contribute to B-cell survival and PB generation by producing BAFF/APRIL and interleukin-21.23 We found splenic neutrophils localized close to PCs and Gr1+Ly6G+ cells as the major source of BAFF. However, the appearance of a Ly6G− subset with strong BAFF expression after anti-Ly6G injection prevented the assessment of PC survival in the context of neutrophil-depleting treatments. Spl-PCs reside in the red pulp at the border of the T-cell zone, but 20% to 30% of them interacted with CD4+ T cells. Such interactions have been previously reported in xenochimeric mice, and we observed them as well in human spleens of ITP patients.24,25 It remains to be assessed whether the role of CD4+ T cells in PC survival is mediated by cell-to-cell contact or by the local production of prosurvival cytokines.24,26 Recently, a subset of T follicular cells has been shown to provide BAFF in GCs.26 However, although anti-CD4 antibodies given alone had little or no effect, the combination of CD4+ T-cell depletion with anti-CD20 treatment significantly decreased the number of Spl-PCs. Of note, T cells isolated from anti-CD20–treated mice promoted the survival of PCs in vitro, but not CD4+ T cells primed with BAFF. Thus, modulation or activation of CD4+ T cells during B-cell depletion via another cellular partner could endow them with additional supportive capacities for LLPC maturation.
Collectively, our results in mice provide strong arguments that combining rituximab with anti-BAFF monoclonal antibody may interfere with the BAFF dependence of autoreactive LLPCs in the spleen and consequently avoid resorting to splenectomy for patients with autoimmune cytopenia, a possibility evaluated in a prospective trial (NCT03154385). Furthermore, because BAFF levels have been shown to affect the threshold of B-cell–positive selection, simultaneous anti-CD20 and anti-BAFF treatment could have a dual function by interfering with PC persistence and contributing to the elimination of new naive B cells with autoimmune potential.27-29
The microarray data presented in this article have been submitted to the ArrayExpress database (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-6449.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Espeli for helpful discussions, L. Da Silva, A. Verge de los Aires, and D. Lecoeuche for technical assistance, and A. Diek for contributing to confocal analyses. The authors also thank R. Zoubairi for animal care, C. Cordier for cell sorting, and N. Goudin for confocal microscopy (from, respectively, the animal, cytometry, and imaging core facilities of the Structure Fédérative de Recherche Necker, INSERM US24-CNRS UMS 3633). The authors also thank Cherié Butts (Biogen Idec) and Sarah Brett (GSK) for providing the anti-CD20 and anti-BAFF antibodies, respectively.
The “Development of the Immune System” team was supported by an ANR PRTS 2013 grant (“PC-RITUX”), the Ligue Nationale contre le Cancer (“Equipe labelisée”), the Fondation Princesse Grace, and a European Research Council Advanced Grant (Memo-B). L.-H.T. was supported by a fellowship from the Société de Médecine Interne (Bourse Marcel Simon) and a Poste d'Accueil INSERM. A.R. was supported by the Société de Médecine Interne (Bourse CSL-Behring). E.C. was supported by a Poste d'Accueil INSERM.
Authorship
Contribution: M.M., C.-A.R., J.-C.W., and L.-H.T. designed the experiments; L.-H.T., M.M., A.R., S.L.G., E.C., Z.Z., J.M., and C.B. performed the experiments; N.C. and T.F. performed the bioinformatic analyses; and M.M., L.-H.T., J.-C.W., and C.-A.R. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: M.M. has received research support from GSK. The remaining authors declare no competing financial interests.
Correspondence: Claude-Agnès Reynaud, Institut Necker-Enfants Malades-INSERM U1151-Team 12 “Développement du Système Immunitaire,” Université Paris Descartes, Faculté de Médecine, Paris, France; e-mail: claude-agnes.reynaud@inserm.fr; and Jean-Claude Weill, Institut Necker-Enfants Malades-INSERM U1151-Team 12 “Développement du Systéme Immunitaire,” Université Paris Descartes, Faculté de Médecine, Paris, France; e-mail: jean-claude.weill@inserm.fr.
References
Author notes
S.L.G. and A.R. contributed equally to this work.

![Figure 1. Gene expression profiling of newly generated and mature splenic EYFP+ PCs from mice. Splenic newly generated EYFP+B220− PCs were sorted 6 days after a third immunization with SRBCs (day 6 [D6]/boost), and splenic mature EYFP+B220− PCs 3 months after the second immunization (M3/controls) (see protocol in supplemental Figure 2A). About 10 000 EYFP+ PCs per mouse were analyzed (D6, n = 3; M3, n = 4). (A) Sorting strategy of mature splenic EYFP+ B220− PCs by use of a 2-step procedure. (B) Total number of EYFP+B220− PCs in controls (M3) or boosted (D6) mice. (C) Heatmap of genes selected from the supervised microarray comparison of newly generated (D6) vs mature PCs (M3) in the mouse spleen (223 genes with fold change in expression >4 or <0.25, P < .05). The heatmap is based on normalized log2 intensities that were probe-wise mean-centered across the 2 compared conditions. Columns represent individual samples, and rows represent specific gene probes, with upregulated genes in red and downregulated genes in blue. Genes selected for the single-cell analysis are highlighted. (D) Functional analysis of the 174 upregulated genes in mature vs newly generated PCs (more than fourfold upregulated; P < .05). The number of genes in each functional category is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f1.jpeg?Expires=1769082321&Signature=QpjEYC0ErpbAw0QyEqCVgLDR0fB3VRpsFHhzH9eaACbJTR6auSb1Mu4hRVEgeBCG5jLTDyDYDv9eKWOg2ridRYTYv~rbyXMnbmSZSfDbZoERFJbjDEElJ~Inpnl5-Bx8Z500dubBY8bFubDve5rQaZHDvEfDS8mDYFa05yViO~ZO2CNEyA2xF1aa7jNnVX7--lbFMA1NPkz7mA4VQI2siWaVKs~0M6XF1w9AQR7no0pfhF7~fOOXsSPUYoxDU3BbZN8G1TXH7cNPemhIx4dyXGBDZxqnvF0pHityOLc8E6oDV3nBu11GFjdcGupgjfMAVXv3dhWieTDbdNONZHM9gA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Maturation of splenic EYFP+ PCs on B-cell depletion. (A) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on PBs, Spl-PCs, anti-CD20 PCs, and BM-PCs using 18 genes (PB genes: Mki67, Chek1, Bub1, Aurka, Rrm2b, Ccnd2, Bcl2l11, and Cxcr3; LLPC genes: Tesc, Klf6, Tnfaip3, Bcl2, Runx2, Tnfrsf17, Nfkb2, Tox2, Tmem176b, and Mt2) discriminating PBs from LLPCs. Analysis was restricted to cells expressing B2m and the Prdm1 PC master gene. Columns represent individual gene probes, and rows represent single cells, with color relative to the cycle threshold expression value (scale bar at bottom). (B) Principal component analysis of 441 individual cells analyzed for the expression of 18 diagnostic genes. Each dot represents a single cell: gray, splenic ASCs generated 3 days after a third immunization (boost); blue, PCs 3 months after immunization (Spl-PCs); orange, PCs at the nadir of B-cell depletion (anti-CD20 PCs); green, BM-PCs 3 months after the second immunization (same animals as Spl-PCs). Multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array involved 140 cells per group (C) Percentage of individual cells expressing n diagnostic PC genes in each population (boost [gray], Spl-PCs [blue], anti-CD20 PCs [orange], BM-PCs [green]). (D) Cumulative percentage of cells expressing n or more PC genes in each group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f3.jpeg?Expires=1769082321&Signature=cAaTQPpanw0P2uw7ZfmpRQr0QNci~jF-ukYmksE6d32W6zKBLczB0Ik1uXfSVlau1fxlVsIXJ9iDpPabnOk4xMpPcZHTRhvQrpgktQKFniIkdG9LqQegrKz1ekAFSU7059R7WxgrZ8dsTqIB3Q0mP8IhXRqjlXa5ehHgUB7a916biMay109ZCuOXZd7ppnNN4GXZIhruXFZ5N7D-uqPcB6GC9OfHy2INY5Q7m~aT-ADw26HR5nRmw3ekV7txPjR~3AZJu55kYWogsqqm6hrFWPQkhAkRhUHLkS3M2Ihw2eZq8NG0pOrHtZIjJIJMbVc5kAytbrCGHmI9h0tGdXsmMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Splenic LLPCs depend on BAFF in the context of B-cell depletion in NZB/W mice. (A) Gating strategy for sorting ASCs from NZB/W mice for single-cell analysis. ASCs from 27-week-old NZB/W mice were sorted after dump-staining with CD3/CD4/Gr1/Cd11b by a 2-step procedure and separated into CD138highB220− and CD138highB220low fractions. ASCs were collected from the spleen (CD138highB220−, CD138highB220low) and bone marrow (CD138highB220−) from 2 untreated and 4 anti-CD20–treated mice at the nadir of B-cell depletion. (B) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on splenic CD138highB220low and CD138highB220− populations using diagnostic genes allowing the discrimination of PBs from LLPCs as in Figure 3A. (C) Percentage of individual cells expressing n PC genes in each population (CD138highB220low [gray], CD138highB220− cells [blue], CD138highB220− anti-CD20 PCs [purple], BM-PCs [green]). (D) Cumulative percentage of individual cells expressing n or more PC genes in each group. (E) Comparison at the nadir of B-cell depletion of CD138highB220− cell numbers in the spleens of mice. Analysis from 2 independent experiments. Significant differences were estimated by 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001; **** P < .0001). Mean values are indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f7.jpeg?Expires=1769082321&Signature=orbu961Ans1n5dnLqBDu9IFPJzEl-4elJfIKvgzNgmGagU5Xa3JqUo0D2XmBJjzMC3QuPcr74WufpOK68dOsKS7K0HIGV0SNEbNkHZgXB7jbwpCL~7LOvJTl~vOs8fxhpYV1l1lNmtYrMfaCmZ-bPO5mEhBqQ6BOIuh0gcX6Gar2ljuXKT43WtwEdSD5YHuTNk83oaD9A8YwZhAqES286Wef8BrqsqycktXOjSSBNA~5IDqvBGx8RLikm87uEzIf0Ek9aXGJraQeSMbNyiLdbcAt53xBNfWj5N03bH4Ldj-3Xg8UJ7yDAeEYkHS7zQ5rIcHkXBZPAm5WiIGL8r~C8g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


![Figure 1. Gene expression profiling of newly generated and mature splenic EYFP+ PCs from mice. Splenic newly generated EYFP+B220− PCs were sorted 6 days after a third immunization with SRBCs (day 6 [D6]/boost), and splenic mature EYFP+B220− PCs 3 months after the second immunization (M3/controls) (see protocol in supplemental Figure 2A). About 10 000 EYFP+ PCs per mouse were analyzed (D6, n = 3; M3, n = 4). (A) Sorting strategy of mature splenic EYFP+ B220− PCs by use of a 2-step procedure. (B) Total number of EYFP+B220− PCs in controls (M3) or boosted (D6) mice. (C) Heatmap of genes selected from the supervised microarray comparison of newly generated (D6) vs mature PCs (M3) in the mouse spleen (223 genes with fold change in expression >4 or <0.25, P < .05). The heatmap is based on normalized log2 intensities that were probe-wise mean-centered across the 2 compared conditions. Columns represent individual samples, and rows represent specific gene probes, with upregulated genes in red and downregulated genes in blue. Genes selected for the single-cell analysis are highlighted. (D) Functional analysis of the 174 upregulated genes in mature vs newly generated PCs (more than fourfold upregulated; P < .05). The number of genes in each functional category is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f1.jpeg?Expires=1769183620&Signature=cbOaZiniybQpsOyDofaUI~nD6GRd-oc4MjKMDawcN7z7o1c~7PuXlL~GPHLbEVSP8KOrI059JlLujyqCcyNVtjngUvlSdsaxKVf~22m59Yg7oeIgkYjITEHtp2uFVLNwtwh3grMVeLkbRbp7oT2xDANN0snt6NTrZ44afx5Eorc9IwwLtG~ykUL-PBs2byKXCFd7e4zpKA1xhu8fxc-7uKEGBGFZ~cjmuWZbVu6eiwhySSEAB3TrfGqyg913E2rMEDmGP8sH8aPwG2KS1gVgRtPMQ9JMmLw-IKfPQ84v8NI7uEo~CssO1cRa0s06i62UQ17SJT1CFRzH44T4Jmjoqg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 3. Maturation of splenic EYFP+ PCs on B-cell depletion. (A) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on PBs, Spl-PCs, anti-CD20 PCs, and BM-PCs using 18 genes (PB genes: Mki67, Chek1, Bub1, Aurka, Rrm2b, Ccnd2, Bcl2l11, and Cxcr3; LLPC genes: Tesc, Klf6, Tnfaip3, Bcl2, Runx2, Tnfrsf17, Nfkb2, Tox2, Tmem176b, and Mt2) discriminating PBs from LLPCs. Analysis was restricted to cells expressing B2m and the Prdm1 PC master gene. Columns represent individual gene probes, and rows represent single cells, with color relative to the cycle threshold expression value (scale bar at bottom). (B) Principal component analysis of 441 individual cells analyzed for the expression of 18 diagnostic genes. Each dot represents a single cell: gray, splenic ASCs generated 3 days after a third immunization (boost); blue, PCs 3 months after immunization (Spl-PCs); orange, PCs at the nadir of B-cell depletion (anti-CD20 PCs); green, BM-PCs 3 months after the second immunization (same animals as Spl-PCs). Multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array involved 140 cells per group (C) Percentage of individual cells expressing n diagnostic PC genes in each population (boost [gray], Spl-PCs [blue], anti-CD20 PCs [orange], BM-PCs [green]). (D) Cumulative percentage of cells expressing n or more PC genes in each group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f3.jpeg?Expires=1769183620&Signature=gQQwwRqHRDK6eskgu9t40VhI-uNMK6duodG~FgIfqk8p-l7Mk88kUxSu6gG1myAdkw~NETHxh1vOK0sr5VzQsMRS6Uibm0Y6GLs~Sn3QebtMb3EbuYLD7ybSd~uTOvN6H3UHmqp9HhhgJ956A7tb9JPtAbDtXDAlNra13h01YpyUdAq357VSBI98j3OSncdaI15j-gIDPW7IMOYcq4c0afczivPO7uN0WRZYDZQNCSP1gHTiGnhwu-zNmyhedXh1jGP1ThGSA4fJ1lEsUEiCK6NYVt7zugQp1FDt74ZNWK08WrMuUOLxWcvYkGGkXkiMScendJw84-tnBicfFEnuzA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Splenic LLPCs depend on BAFF in the context of B-cell depletion in NZB/W mice. (A) Gating strategy for sorting ASCs from NZB/W mice for single-cell analysis. ASCs from 27-week-old NZB/W mice were sorted after dump-staining with CD3/CD4/Gr1/Cd11b by a 2-step procedure and separated into CD138highB220− and CD138highB220low fractions. ASCs were collected from the spleen (CD138highB220−, CD138highB220low) and bone marrow (CD138highB220−) from 2 untreated and 4 anti-CD20–treated mice at the nadir of B-cell depletion. (B) Heatmap representation of multiplex single-cell RT-PCR performed with the Fluidigm Dynamic Array on splenic CD138highB220low and CD138highB220− populations using diagnostic genes allowing the discrimination of PBs from LLPCs as in Figure 3A. (C) Percentage of individual cells expressing n PC genes in each population (CD138highB220low [gray], CD138highB220− cells [blue], CD138highB220− anti-CD20 PCs [purple], BM-PCs [green]). (D) Cumulative percentage of individual cells expressing n or more PC genes in each group. (E) Comparison at the nadir of B-cell depletion of CD138highB220− cell numbers in the spleens of mice. Analysis from 2 independent experiments. Significant differences were estimated by 1-way analysis of variance, Dunn test for multiple comparisons (* P < .05; *** P < .001; **** P < .0001). Mean values are indicated.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/14/10.1182_blood-2017-06-789578/4/m_blood789578f7.jpeg?Expires=1769183620&Signature=CGtd-MtFb5-dnY8BMdIt-z67YA8zxwPI8SiPPdwWnZEF9LHfXZYgxnzDdiylYZllmKbAQbizzUiX2ugKblR4YxJHCahGzPnxRGgwTmezqtRaIRyrhqLcA--gt6TDxinjyKFyn0epiq8-MicMLV3r7JcBhV5E6hz5LamXOFJD3KCPYEi4tvma8O65c2vj1fAnw1oO4UPlT~6GGcJcmbFrXjXJp-C~fPVs4~pnco0U4Fn9lbaGm~cm976pqCn0NXu1TlZRdVDoQULCXrWxOXwTHUACaQrvQMlLOGMGPY-oel9dh~A45Gub-L-yaUQe0~AslgbKGN7WD-pjVaJPVCPlIQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)