Key Points
CRISPR/Cas9-mediated disruption of the β-globin locus architecture reactivates fetal γ-globin expression in adult erythroblasts.
Fetal γ-globin reactivation and sickle β-globin downregulation leads to the amelioration of the SCD cell phenotype.
Abstract
Naturally occurring, large deletions in the β-globin locus result in hereditary persistence of fetal hemoglobin, a condition that mitigates the clinical severity of sickle cell disease (SCD) and β-thalassemia. We designed a clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) strategy to disrupt a 13.6-kb genomic region encompassing the δ- and β-globin genes and a putative γ-δ intergenic fetal hemoglobin (HbF) silencer. Disruption of just the putative HbF silencer results in a mild increase in γ-globin expression, whereas deletion or inversion of a 13.6-kb region causes a robust reactivation of HbF synthesis in adult erythroblasts that is associated with epigenetic modifications and changes in chromatin contacts within the β-globin locus. In primary SCD patient-derived hematopoietic stem/progenitor cells, targeting the 13.6-kb region results in a high proportion of γ-globin expression in erythroblasts, increased HbF synthesis, and amelioration of the sickling cell phenotype. Overall, this study provides clues for a potential CRISPR/Cas9 genome editing approach to the therapy of β-hemoglobinopathies.
Introduction
β-Thalassemia and sickle cell disease (SCD) are severe anemias caused by mutations in the β-globin gene cluster. In β-thalassemia, the reduced production of adult β-chains causes α-globin precipitation, ineffective erythropoiesis, and insufficiently hemoglobinized red blood cells (RBCs). In SCD, the β6Glu→Val substitution leads to Hb polymerization and RBC sickling, which is responsible for vaso-occlusive crises, hemolytic anemia, and organ damage. Current treatment of SCD and β-thalassemia involves regular RBC transfusion, which is associated with significant side effects, such as iron overload and organ damage. The only definitive cure for β-hemoglobinopathies is allogeneic hematopoietic stem cell (HSC) transplantation from HLA-matched sibling donors, which is available only to a fraction of patients.1-4 Transplantation of autologous, genetically corrected HSCs is an attractive therapeutic alternative for patients lacking a compatible intrafamilial donor.5,6
The clinical course of β-hemoglobinopathies is ameliorated by elevated levels of fetal γ-globin, which reduces globin chain imbalance in β-thalassemias and exerts a potent antisickling effect in SCD. Naturally occurring large deletions encompassing the β- and δ-globin genes result in a congenital increase of fetal hemoglobin (HbF) expression known as hereditary persistence of HbF (HPFH), which ameliorates both thalassemic and SCD phenotypes. Large HPFH deletions are thought to eliminate HbF inhibitory sequences or juxtapose the γ-globin promoters to remote enhancer regions.7 Several studies demonstrated that BCL11A, a transcriptional repressor, is required to maintain silencing of HbF expression.8,9 BCL11A interacts with GATA1, FOG1, SOX6, and the NuRD repressor complex and occupies critical sites within the β-globin gene cluster, including sequences specifically deleted in HPFH individuals.10,11 These observations provide a strong rationale for developing gene therapy and genome editing12-19 approaches to the treatment of β-hemoglobinopathies aimed at inducing a β-to-γ globin reverse switching.
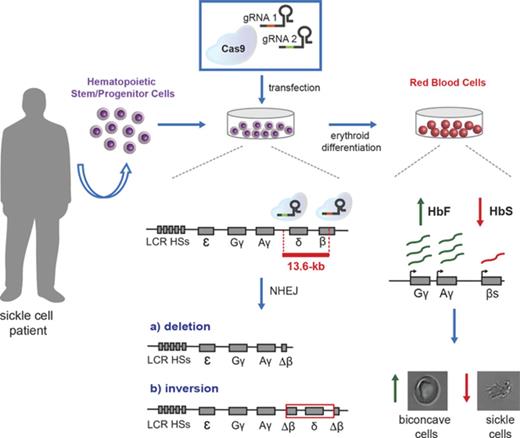
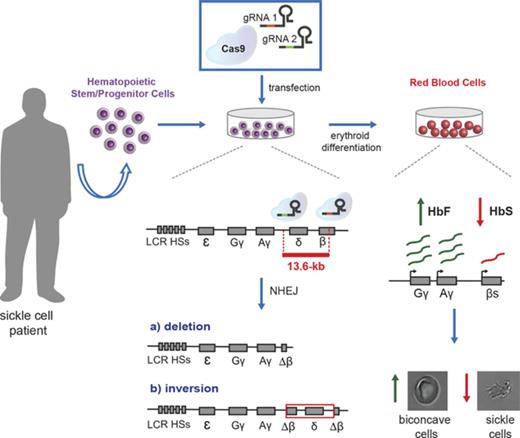
Here, we integrated transcription factor binding site analysis and HPFH genetic data to define cis-regulatory elements in the β-globin locus involved in γ-globin gene silencing. We designed clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) (CRISPR/Cas9) nucleases disrupting (1) a potential γ-δ intergenic HbF silencer containing a putative BCL11A binding site10 ; (2) the shortest deletion associated with elevated HbF levels in β-thalassemic patients (“Corfu” deletion), encompassing the putative γ-δ intergenic HbF silencer20-22 ; and (3) an extended, 13.6-kb genomic region including the δ- and β-globin genes and the putative intergenic HbF silencer. Disrupting the 13.6-kb region led to a robust HbF reactivation and a concomitant reduction in β-globin expression in an adult erythroid cell line23 and in healthy donor and SCD hematopoietic stem/progenitor cell (HSPC)-derived erythroblasts.
Methods
Cell culture
HUDEP-2 cells were cultured and differentiated, as described by Canver et al.14 Genome-edited HUDEP-2 bulk populations were cloned by limiting dilution and clones harboring deletion or inversion were screened by polymerase chain reaction (PCR) and Droplet Digital PCR (ddPCR). Healthy donor HSPCs were differentiated toward the erythroid lineage as previously described.8 SCD HSPCs were terminally differentiated in RBCs using a 3-phase erythroid culture system.24
Cell transfection
Cells were transfected with 4 μg of a Cas9-GFP expressing plasmid and 0.8 to 1.6 μg of each guide RNA (gRNA)–containing vector using Nucleofector I (Lonza). We used AMAXA Cell Line Nucleofector Kit V (VCA-1003) for K562 and HUDEP-2 (T16 and L-29 programs) and AMAXA Human CD34 Cell Nucleofector Kit (VPA-1003) for HSPCs (U-08 program). GFP+ cells were sorted using SH800 Cell Sorter (Sony Biotechnology).
3C and ChIP assays
Sickling assay
In vitro–generated SCD RBCs were exposed to an oxygen-deprived atmosphere (0% O2), and the time course of sickling was monitored in real time by video microscopy for 1 hour, capturing images every 5 minutes using the AxioObserver Z1 microscope (Zeiss) and a 40× objective. Images of the same fields were taken throughout all stages and processed with ImageJ to determine the percentage of sickled RBCs per field of acquisition in the total RBC population.
Results
Identification of potential therapeutic targets for HbF reactivation in the β-globin locus
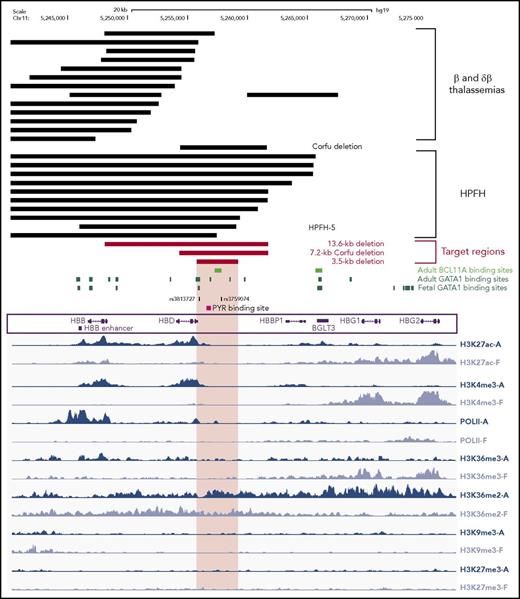
In HPFH individuals, genomic deletions encompassing the β- and δ-globin genes lead to therapeutic HbF levels. In contrast, in deletional β- and δβ-thalassemias, HbF synthesis is not sufficient to allow a functional RBC maturation. An exhaustive comparison of large (>7 kb) genomic deletions associated with either HPFH or β0- and δβ0-thalassemias identified a 3.5-kb γ-δ intergenic region absent in 10 out of 12 HPFH deletions that represents a potential cis-regulatory element involved in HbF silencing (Figure 1; supplemental Figure 1, available on the Blood Web site). By reanalyzing publicly available ChIP-sequencing data,27 we identified in this region binding sites for the erythroid-specific master transcription factor GATA1 in adult, but not fetal, primary erythroblasts (Figure 1). The 3.5-kb region contains single-nucleotide polymorphisms associated with high HbF levels,28,29 binding sites for the chromatin remodeling PYR complex, which may be involved in HbF silencing,30 and a putative BCL11A binding site10,31 (Figure 1). The same region is devoid of histone modifications associated with either active (H3K27ac and H3K36me3) or repressive (H3K27me3 and H3K9me3) chromatin states27 in both fetal and adult erythroblasts (Figure 1), while it is enriched in H3K36 dimethylation (H3K36me2), a histone modification that may generate a repressive chromatin environment.27,32,33 H3K36me2 is in fact enriched in fetal globin genes in adult erythroblasts and in adult globin genes in fetal erythroblasts (Figure 1). The 3.5-kb γ-δ intergenic region is included in the 7.2-kb Corfu deletion, the minimal deletion resulting in HbF elevation in β-thalassemic patients34 (Figure 1). In contrast, 13 out of 19 deletions causing β0- and δβ0-thalassemias do not include the 3.5-kb region and/or the putative BCL11A binding site (Figure 1; supplemental Figure 1). Of the 6 δβ0-thalassemia deletions removing the 3.5-kb region, 2 (Thai35 and black36 δβ0-thalassemias) are associated with extremely high (25%) HbF levels that significantly improve β-thalassemic and SCD phenotypes, while the remaining ones are associated with a mild thalassemic phenotype in heterozygous and homozygous patients10,37-40 (supplemental Figure 1).
Integration of mutational and epigenetic analyses of the β-globin locus. Genomic deletions mapped in thalassemic patients and HPFH individuals are indicated with black bars (top). We report the 10 HPFH mutations removing the 3.5-kb region and the 13 β-thalassemia-associated deletions, which do not include this region. The 13.6-kb, the 7.2-kb Corfu and the 3.5-kb target regions are depicted as red bars. The 3.5-kb region contains a putative BCL11A binding site and several GATA1 binding sites (retrieved from Xu et al,27,31 and Jawaid et al45), single-nucleotide polymorphisms associated with high HbF levels, and a 250-bp polypyrimidine-rich sequence targeted by the PYR complex. We analyzed the histone modification pattern of the β-globin locus in human adult and fetal erythroblasts27 (in blue and lavender, respectively; bottom). Epigenetic modifications typical of active chromatin regions, such as H3K27 acetylation (H3K27ac), H3K4 trimethylation (H3K4me3), H3K36 trimethylation (H3K36me3), and RNA polymerase II binding (PolII), mark the β- and δ-globin and the γ-globin genes in adult and fetal erythroblasts, respectively. Typical repressive chromatin markers (H3K9 trimethylation, H3K9me3 and H3K27 trimethylation, H3K27me3) were absent in the β-globin locus of both adult and fetal erythroblasts. The 3.5-kb target region, as well as inactive γ-globin genes, were preferentially enriched in H3K36 dimethylation (H3K36me2) in adult cells. HBB, β-globin gene; HBD, δ-globin gene; HBBP1, β-globin pseudogene 1; BGLT3, β-globin locus transcript 3 gene; HBG1, Aγ-globin gene; HBG2, Gγ-globin gene. The enhancer located 3′ to the poly(A) site of the β-globin gene is indicated as HBB enhancer.
Integration of mutational and epigenetic analyses of the β-globin locus. Genomic deletions mapped in thalassemic patients and HPFH individuals are indicated with black bars (top). We report the 10 HPFH mutations removing the 3.5-kb region and the 13 β-thalassemia-associated deletions, which do not include this region. The 13.6-kb, the 7.2-kb Corfu and the 3.5-kb target regions are depicted as red bars. The 3.5-kb region contains a putative BCL11A binding site and several GATA1 binding sites (retrieved from Xu et al,27,31 and Jawaid et al45), single-nucleotide polymorphisms associated with high HbF levels, and a 250-bp polypyrimidine-rich sequence targeted by the PYR complex. We analyzed the histone modification pattern of the β-globin locus in human adult and fetal erythroblasts27 (in blue and lavender, respectively; bottom). Epigenetic modifications typical of active chromatin regions, such as H3K27 acetylation (H3K27ac), H3K4 trimethylation (H3K4me3), H3K36 trimethylation (H3K36me3), and RNA polymerase II binding (PolII), mark the β- and δ-globin and the γ-globin genes in adult and fetal erythroblasts, respectively. Typical repressive chromatin markers (H3K9 trimethylation, H3K9me3 and H3K27 trimethylation, H3K27me3) were absent in the β-globin locus of both adult and fetal erythroblasts. The 3.5-kb target region, as well as inactive γ-globin genes, were preferentially enriched in H3K36 dimethylation (H3K36me2) in adult cells. HBB, β-globin gene; HBD, δ-globin gene; HBBP1, β-globin pseudogene 1; BGLT3, β-globin locus transcript 3 gene; HBG1, Aγ-globin gene; HBG2, Gγ-globin gene. The enhancer located 3′ to the poly(A) site of the β-globin gene is indicated as HBB enhancer.
This analysis suggests that the 3.5-kb γ-δ intergenic region and the 7.2-kb Corfu region represent potential therapeutic targets to achieve HbF reactivation by a targeted deletion approach (Figure 1). In addition, we designed a larger, 13.6-kb deletion starting from the 5′ breakpoint of the Corfu deletion and extending further 3′ to include the promoter and first exon of the β-globin gene, similar to the naturally occurring 12.9-kb HPFH-5 deletion41 (Figure 1; supplemental Figure 1). Compared with the HPFH-5 deletion, the 13.6-kb deletion leaves intact the 3′ and intronic enhancers of the β-globin gene that, upon juxtaposition to the γ-globin genes, may further enhance their expression (Figure 1; supplemental Figure 1). In SCD patients, the 13.6-kb deletion would simultaneously upregulate γ-globin synthesis and inactivate the βS-globin gene.
Disruption of the 13.6-kb region reactivates fetal γ-globin expression in an adult erythroblastic cell line
To generate the deletion of the target regions, we designed 18-bp single gRNAs42 recognizing the 5′ and 3′ ends of the selected genomic loci (supplemental Figure 2A). For each target site, 3 to 5 gRNAs (supplemental Table 1) were individually delivered, together with a Cas9-GFP fusion protein, in K562 erythroid cells by plasmid transfection. We selected efficient gRNAs able to generate insertions and deletions (InDels) in the target sites with an efficiency of ≥55% in Cas9-GFP+ cells (supplemental Figure 2B-C). Pairs of the selected gRNAs successfully generated 3.5-, 7.2-, and 13.6-kb deletions in up to 31% of the alleles and inversions of the same regions at lower frequencies (supplemental Figure 2D). At the remaining loci, we measured up to 68% of small InDels at both 5′ and 3′ gRNA target sites without excision or inversion of the intervening sequence (supplemental Figure 2D).
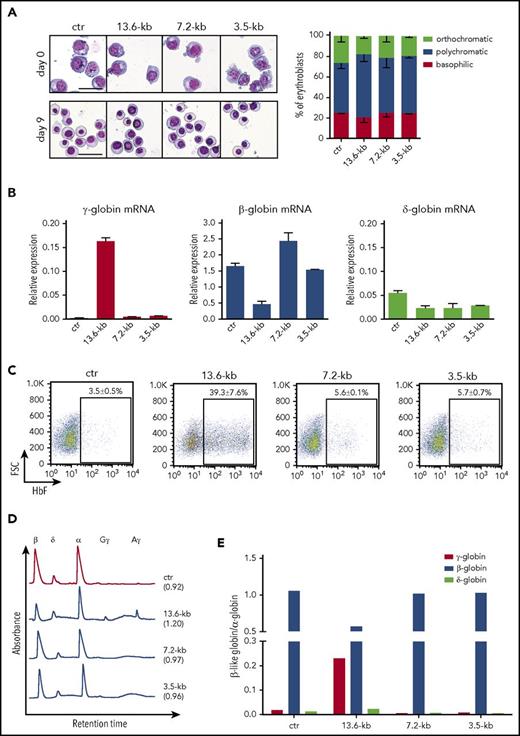
To evaluate HbF reactivation upon CRISPR/Cas9-mediated disruption of the targeted regions, we transfected plasmids encoding Cas9-GFP and gRNA pairs into HUDEP-2 erythroid cells expressing mainly adult β-globin.23 The deletion and inversion frequency was 25% ± 8% and 39% ± 3%, respectively, for the 13.6-kb region, 27% ± 6% and 21% ± 11% for the 7.2-kb region, and 18% ± 3% and 6% ± 2% for the 3.5-kb region. Minimal InDel frequency was observed at the top-predicted off-target sites (supplemental Table 2). The frequency of deletion/inversion events did not change upon terminal differentiation, thus excluding negative selection of genome-edited cells during erythroid maturation (supplemental Figure 3A). Morphological analysis of cell cultures revealed no differences between genome-edited and control cells (Figure 2A), indicating that erythroid differentiation is not affected by disruption of the β-globin locus. The expression of transcription factors involved in hemoglobin switching and HbF silencing did not change in control and treated samples (supplemental Figure 3B). Terminally differentiated HUDEP-2 bulk populations harboring deletion and inversion of the 13.6-kb showed a dramatic increase in γ-globin messenger RNA (mRNA) levels and concomitant decrease in β- and δ-globin expression (Figure 2B). Fluorescence activated cell sorting (FACS) analysis showed a substantial increase in the proportion of F-cells compared with control samples (Figure 2C). Reverse-phase high-performance liquid chromatography (RP-HPLC) analysis confirmed reactivation of Aγ and Gγ-globin chains and a decrease in β-globin expression (Figure 2D-E). Interestingly, the extent of genome editing in the 13.6-kb region was positively correlated with the increase in the percentage of F-cells and HbF production (supplemental Figure 4).
Targeting of a 13.6-kb genomic region in the β-globin locus reactivates γ-globin expression in the adult HUDEP-2 erythroid cell line. HUDEP-2 cells were transfected with plasmids carrying Cas9-GFP and gRNA pairs targeting the 13.6-kb, 7.2-kb, and 3.5-kb regions. Cells treated only with Cas9-GFP plasmid were used as control (ctr). GFP+ cells were FACS-sorted and differentiated into mature erythroblasts. (A) Representative images of May-Grünwald-Giemsa–stained undifferentiated (day 0) and differentiated (day 9) cultures. Original magnification ×20. Scale bars, 50 µm (left). Bars indicate the percentage cell number for each erythroblast population after differential counting (right). Similar proportions of the different erythroid precursors were observed in control and genome-edited cultures. (B) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin mRNA levels in differentiated samples. Results were normalized to α-globin. Error bars denote standard deviation (SD). (C) Representative FACS analyses of HbF+ cells (F-cells). Data are expressed as mean ± standard error of the mean (SEM) of 3 experiments. (D) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in differentiated HUDEP-2 samples. The ratio of α chains to non–α chains is indicated in brackets. (E) Quantification of γ (Aγ+Gγ)-globin and β-globin protein levels, as assessed by RP-HPLC. β-Like globin expression was normalized to α-globin. Targeting the 13.6-kb region, but not the 3.5-kb putative HbF silencer and the 7.2-kb region, reduced β-globin chain levels and strongly increased γ-globin chain expression. δ-Globin protein levels were decreased only in 7.2-kb and 3.5-kb genome-edited samples.
Targeting of a 13.6-kb genomic region in the β-globin locus reactivates γ-globin expression in the adult HUDEP-2 erythroid cell line. HUDEP-2 cells were transfected with plasmids carrying Cas9-GFP and gRNA pairs targeting the 13.6-kb, 7.2-kb, and 3.5-kb regions. Cells treated only with Cas9-GFP plasmid were used as control (ctr). GFP+ cells were FACS-sorted and differentiated into mature erythroblasts. (A) Representative images of May-Grünwald-Giemsa–stained undifferentiated (day 0) and differentiated (day 9) cultures. Original magnification ×20. Scale bars, 50 µm (left). Bars indicate the percentage cell number for each erythroblast population after differential counting (right). Similar proportions of the different erythroid precursors were observed in control and genome-edited cultures. (B) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin mRNA levels in differentiated samples. Results were normalized to α-globin. Error bars denote standard deviation (SD). (C) Representative FACS analyses of HbF+ cells (F-cells). Data are expressed as mean ± standard error of the mean (SEM) of 3 experiments. (D) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in differentiated HUDEP-2 samples. The ratio of α chains to non–α chains is indicated in brackets. (E) Quantification of γ (Aγ+Gγ)-globin and β-globin protein levels, as assessed by RP-HPLC. β-Like globin expression was normalized to α-globin. Targeting the 13.6-kb region, but not the 3.5-kb putative HbF silencer and the 7.2-kb region, reduced β-globin chain levels and strongly increased γ-globin chain expression. δ-Globin protein levels were decreased only in 7.2-kb and 3.5-kb genome-edited samples.
In contrast, deletion and inversion of the Corfu region or of the 3.5-kb putative HbF silencer did not significantly increase γ-globin expression (Figure 2B-D) in differentiated bulk populations of HUDEP-2 cells. These results were confirmed in HUDEP-2 clones harboring biallelic deletions, which showed no increase in primary and mature γ-globin gene transcripts and HbF accumulation (supplemental Figure 5A-C).
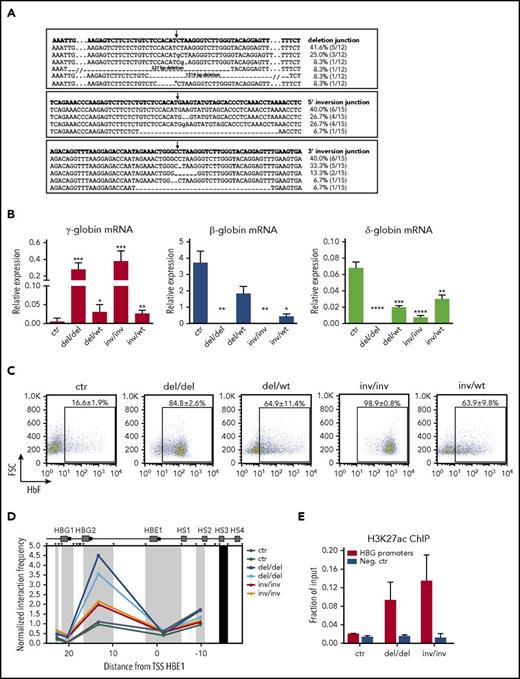
To correlate HbF reactivation with the specific genomic modifications of the 13.6-kb region, we generated clones harboring biallelic or monoallelic 13.6-kb deletions (del/del, del/wt) or inversions (inv/inv and inv/wt). Sequencing of the targeted region showed in the vast majority of the cases the expected repaired junctions in both deleted and inverted alleles, with small InDels (Figure 3A). Erythroid differentiation was not impaired in genome-edited clones compared with control cells (supplemental Figure 6A). We observed increased γ-globin expression levels and HbF production in clones harboring biallelic deletions or inversions, whereas monoallelic deletion/inversion of the 13.6-kb region determined a mild reactivation of HbF (Figure 3B-C). As expected, β-globin expression was not detected in biallelic clones and decreased in monoallelic clones (Figure 3B). δ-Globin mRNA was absent in clones harboring biallelic deletions and significantly reduced in inv/inv, del/wt, and inv/wt clones (Figure 3B).
HbF reactivation occurs predominantly in HUDEP-2 clones harboring biallelic rearrangements of the 13.6-kb target region. Control and genome-edited bulk populations were cloned by limiting dilution to isolate cells harboring biallelic or monoallelic modifications of the 13.6-kb target region. We selected and differentiated 11 control (ctr), 4 biallelic deleted (del/del), 4 mono-allelic deleted (del/wt), 5 biallelic inverted (inv/inv), and 5 monoallelic inverted (inv/wt) clones. (A) Sanger sequencing of deletion and inversion junctions in genome-edited clones. Top rows show the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In 2 alleles, we detected larger deletions (14.1 and 14.9 kb) removing 527 and 1319 bp upstream and downstream of the 13.6-kb region, respectively. In 1 allele, we detected a shorter deletion (11.6 kb), leaving 2001 bp at the 3′ end of the 13.6-kb region (*). No difference in HbF expression was observed among these clones regardless of the deletion. The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes and dots represent deleted and hidden nucleotides, respectively. Inserted nucleotides are displayed in lowercase. (B) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in differentiated clones. All samples were normalized to α-globin. Error bars represent SEM. ****P < .0001, ***P < .001, **P < .01, *P < .05 (unpaired 2-tailed Student t test vs control). (C) Representative FACS plots showing the percentage of F-cells in differentiated HUDEP-2 clones. The proportion of HbF+ cells was significantly higher in genome-edited vs control clones (P < .0001). Data are displayed as mean ± SEM. Clones harboring a biallelic inversion of the 13.6-kb regions tend to show higher γ-globin expression at both mRNA and protein levels than clones harboring a biallelic deletion. (D) 3C analysis of chromatin interactions between the LCR and the γ-globin promoters in differentiated clones (2 controls, 2 del/del, and 2 inv/inv clones). A higher interaction frequency was observed in clones harboring a biallelic rearrangement, as compared with control clones. The interaction frequencies were normalized to the cross-linking frequency in the ERCC3 locus. The hypersensitive sites of the LCR are indicated (HS1, HS2, HS3, and HS4). We used as anchor a genomic fragment containing HS3 (black rectangle). HindIII restriction sites are depicted as black triangles. Black circles indicate the β-like globin promoters. Distances on x-axis are in kilobases counting from the transcription start site (TSS) of the HBE1 gene. (E) Analysis of H3K27 acetylation at γ-globin promoters in differentiated clones (2 controls, 2 del/del, and 2 inv/inv clones). γ-Globin promoters were highly enriched in H3K27ac in del/del and inv/inv clones. The DEFB122 genomic region served as a negative control (Neg. ctr). Error bars indicate SD.
HbF reactivation occurs predominantly in HUDEP-2 clones harboring biallelic rearrangements of the 13.6-kb target region. Control and genome-edited bulk populations were cloned by limiting dilution to isolate cells harboring biallelic or monoallelic modifications of the 13.6-kb target region. We selected and differentiated 11 control (ctr), 4 biallelic deleted (del/del), 4 mono-allelic deleted (del/wt), 5 biallelic inverted (inv/inv), and 5 monoallelic inverted (inv/wt) clones. (A) Sanger sequencing of deletion and inversion junctions in genome-edited clones. Top rows show the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In 2 alleles, we detected larger deletions (14.1 and 14.9 kb) removing 527 and 1319 bp upstream and downstream of the 13.6-kb region, respectively. In 1 allele, we detected a shorter deletion (11.6 kb), leaving 2001 bp at the 3′ end of the 13.6-kb region (*). No difference in HbF expression was observed among these clones regardless of the deletion. The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes and dots represent deleted and hidden nucleotides, respectively. Inserted nucleotides are displayed in lowercase. (B) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in differentiated clones. All samples were normalized to α-globin. Error bars represent SEM. ****P < .0001, ***P < .001, **P < .01, *P < .05 (unpaired 2-tailed Student t test vs control). (C) Representative FACS plots showing the percentage of F-cells in differentiated HUDEP-2 clones. The proportion of HbF+ cells was significantly higher in genome-edited vs control clones (P < .0001). Data are displayed as mean ± SEM. Clones harboring a biallelic inversion of the 13.6-kb regions tend to show higher γ-globin expression at both mRNA and protein levels than clones harboring a biallelic deletion. (D) 3C analysis of chromatin interactions between the LCR and the γ-globin promoters in differentiated clones (2 controls, 2 del/del, and 2 inv/inv clones). A higher interaction frequency was observed in clones harboring a biallelic rearrangement, as compared with control clones. The interaction frequencies were normalized to the cross-linking frequency in the ERCC3 locus. The hypersensitive sites of the LCR are indicated (HS1, HS2, HS3, and HS4). We used as anchor a genomic fragment containing HS3 (black rectangle). HindIII restriction sites are depicted as black triangles. Black circles indicate the β-like globin promoters. Distances on x-axis are in kilobases counting from the transcription start site (TSS) of the HBE1 gene. (E) Analysis of H3K27 acetylation at γ-globin promoters in differentiated clones (2 controls, 2 del/del, and 2 inv/inv clones). γ-Globin promoters were highly enriched in H3K27ac in del/del and inv/inv clones. The DEFB122 genomic region served as a negative control (Neg. ctr). Error bars indicate SD.
3C experiments showed an increased interaction frequency between the locus control region (LCR) and the γ-globin promoters in clones harboring biallelic deletions or inversions (Figure 3D). We observed increased levels of H3K27 acetylation, a marker of active regulatory elements, at the γ-globin promoters in both del/del and inv/inv clones (Figure 3E). 3C experiments indicated a reduced interaction between the LCR and the genomic region containing the adult β- and δ-globin genes in clones harboring the inversion of the 13.6-kb region (supplemental Figure 6B). Interestingly, deletion of the 13.6-kb region led to a high interaction frequency between the Gγ-globin promoter and a genomic region containing the 3′ and intronic enhancers of the β-globin gene, the HBBP1 pseudogene, and the β-globin locus transcript 3 (BGLT3) gene (supplemental Figure 6C).
Induction of γ-globin expression in primary erythroblasts differentiated from genome-edited HSPCs
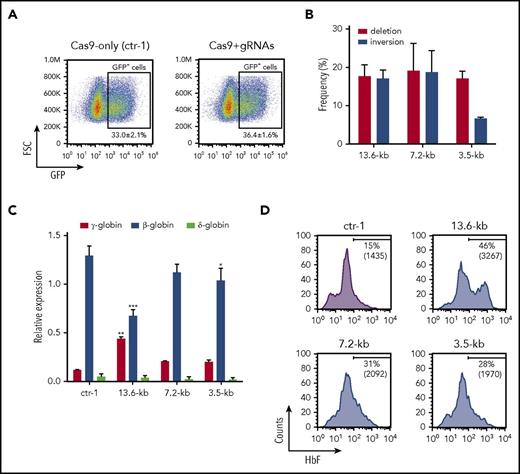
Cas9-GFP and gRNAs targeting the 13.6-, 7.2-, and 3.5-kb regions were delivered by plasmid transfection to granulocyte colony-stimulating factor (G-CSF)–mobilized HSPCs from healthy donors, and GFP+ sorted cells were differentiated in liquid culture toward the erythroid lineage (Figure 4A). In mature erythroblasts, the deletion and inversion frequency was 18% ± 3% and 17% ± 2%, respectively, for the 13.6-kb region, 19% ± 7% and 19% ± 6% for the 7.2-kb region, and 17% ± 2% and 7% ± 1% for the 3.5-kb region (Figure 4B). Quantitative reverse transcription PCR (qRT-PCR) showed an approximately fourfold increase in γ-globin expression and a parallel reduction in β-globin mRNA levels in 13.6-kb genome-edited cells compared with control samples (Figure 4C). A mild (approximately twofold) increase in γ-globin transcript levels was observed in 3.5- and 7.2-kb genome-edited cells (Figure 4C). δ-Globin mRNA levels tended to decrease in all genome-edited samples (Figure 4C). FACS analysis revealed that the percentage of F-cells and their HbF content were increased in all genome-edited samples, with a remarkable and more prominent increment in HbF production in 13.6-kb genome-edited samples (Figure 4D).
Selection of therapeutic targets for γ-globin reactivation in erythroblasts derived from clinically relevant HSPCs. HSPCs derived from healthy donors were transfected with plasmids carrying Cas9-GFP and gRNAs targeting the 13.6-, 7.2-, and 3.5-kb regions. (A) Flow cytometry sorting strategy of control and genome-edited cells. GFP+ HSPCs were FACS-sorted from Cas9-only (ctr-1) and Cas9+gRNAs samples (13.6, 7.2, and 3.5 kb). Data are expressed as mean ± SEM of 7 independent experiments. FSC, forward scatter. (B) Assessment of deletion and inversion efficiency by ddPCR. FACS-sorted control and genome-edited (13.6-kb, 7.2-kb, and 3.5-kb) cells were differentiated in liquid culture toward the erythroid lineage. Data represent the mean ± SD of at least 2 independent experiments. (C) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from GFP+ HSPCs. mRNA levels were normalized to α-globin. γ-Globin expression levels were significantly increased in 13.6-kb genome-edited cells compared with the control sample. A modest increase in γ-globin mRNA was detected in 7.2-kb and 3.5-kb genome-edited samples. A robust and significant β-globin downregulation was observed in 13.6-kb genome-edited erythroblasts compared with control cells. *P < .05, **P < .01; ***P < .001 (2-way analysis of variance, Bonferroni’s multiple comparisons test vs ctr-1). Data represent the mean ± SD of at least 2 independent experiments. (D) Representative FACS histograms showing the increase in both the percentage of F-cells and the median fluorescence intensity (MFI) (in brackets) in mature erythroblasts derived from GFP+ genome-edited HSPCs in comparison with the control sample (Cas9-only cells; ctr-1).
Selection of therapeutic targets for γ-globin reactivation in erythroblasts derived from clinically relevant HSPCs. HSPCs derived from healthy donors were transfected with plasmids carrying Cas9-GFP and gRNAs targeting the 13.6-, 7.2-, and 3.5-kb regions. (A) Flow cytometry sorting strategy of control and genome-edited cells. GFP+ HSPCs were FACS-sorted from Cas9-only (ctr-1) and Cas9+gRNAs samples (13.6, 7.2, and 3.5 kb). Data are expressed as mean ± SEM of 7 independent experiments. FSC, forward scatter. (B) Assessment of deletion and inversion efficiency by ddPCR. FACS-sorted control and genome-edited (13.6-kb, 7.2-kb, and 3.5-kb) cells were differentiated in liquid culture toward the erythroid lineage. Data represent the mean ± SD of at least 2 independent experiments. (C) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from GFP+ HSPCs. mRNA levels were normalized to α-globin. γ-Globin expression levels were significantly increased in 13.6-kb genome-edited cells compared with the control sample. A modest increase in γ-globin mRNA was detected in 7.2-kb and 3.5-kb genome-edited samples. A robust and significant β-globin downregulation was observed in 13.6-kb genome-edited erythroblasts compared with control cells. *P < .05, **P < .01; ***P < .001 (2-way analysis of variance, Bonferroni’s multiple comparisons test vs ctr-1). Data represent the mean ± SD of at least 2 independent experiments. (D) Representative FACS histograms showing the increase in both the percentage of F-cells and the median fluorescence intensity (MFI) (in brackets) in mature erythroblasts derived from GFP+ genome-edited HSPCs in comparison with the control sample (Cas9-only cells; ctr-1).
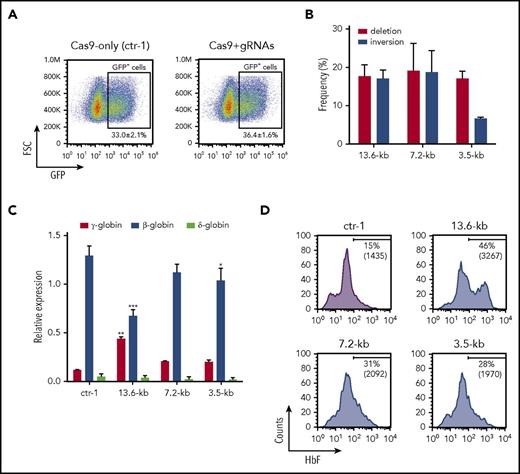
We evaluated precisely the efficiency and efficacy of CRISPR/Cas9-mediated HbF induction upon targeting of the 13.6-kb region in HSPCs derived from healthy donors and SCD patients. We achieved Cas9-GFP expression in 30% to 60% of HSPCs with deletion and inversion efficiency of 7.4% ± 0.3% and 6.8% ± 0.5%, respectively, in unsorted treated cells. In mature erythroblasts derived from FACS-sorted Cas9-GFP+ genome-edited HSPCs, we detected ∼20% of deleted or inverted alleles (Figure 5A). Genome-editing efficiency was similar between pools of erythroid progenitors (BFU-E) and mature erythroblasts generated in liquid culture in both unsorted and sorted genome-edited populations (supplemental Figure 7A; Figure 5A), thus indicating efficient CRISPR/Cas9-mediated genome editing in early hematopoietic progenitors. To estimate precisely the frequency of mono-/biallelic deletion and inversion events, we measured the editing efficiency in single BFU-E. PCR analysis of the targeted region showed a high frequency (∼70%) of clones harboring mono- and biallelic deletions and inversions (Figure 5B). We observed in the majority of the alleles the expected deletion and inversion junctions, with small InDels (Figure 5C). qRT-PCR analysis showed an increase in γ-globin expression and a parallel reduction in β- and δ-globin mRNA levels in genome-edited cells derived from both healthy donor and SCD HSPCs when compared with control samples (Figure 5D). In mature erythroblasts derived from healthy donor GFP+ genome-edited HSPCs, the proportion of F-cells and their HbF content (MFI) were markedly increased (Figure 5E). RP-HPLC analysis revealed a robust increase in γ-globin protein levels (Figure 5F-G) representing up to 53% of the total β-like globin content (Figure 5H), and a twofold reduction of β-globin protein levels (Figure 5F-G). Notably, we did not observe imbalance in α- and β-like globin chain synthesis in genome-edited cells, thus indicating that the reduction of β-globin is compensated by the increased production of γ-globin chains (Figure 5F). On the contrary, β-globin gene knockdown, performed by using a gRNA targeting the exon 1 of the β-globin, induced a modest increase in γ-globin levels, which were insufficient to compensate the reduction of β-globin expression, thus resulting in the imbalance in α- and β-like globin chain synthesis (supplemental Figure 8A) and generation of α-globin aggregates, as observed in mature erythrocytes of β-thalassemic patients43 (supplemental Figure 8B). Erythroid differentiation was not hampered upon CRISPR/Cas9-mediated genome editing, as determined by FACS analysis of erythroid markers (supplemental Figure 8C) and morphological analyses (supplemental Figure 8D). Similarly, HbF upregulation was observed in pools of BFU-E derived from GFP+ genome-edited cells (supplemental Figure 7G-I). Mature erythroblasts derived from unsorted genome-edited HSPCs showed a less pronounced but consistent increase in HbF production, as demonstrated by qRT-PCR, FACS, and RP-HPLC analyses (supplemental Figure 7B-F). Importantly, the percentage of genome-edited alleles was directly correlated to the γ-globin mRNA and protein levels, and the increased percentage of F-cells in mature erythroblasts (P < .05; supplemental Figure 9).
Robust HbF upregulation in HSPC-derived erythroblasts upon genome editing of the 13.6-kb region. (A) Assessment of deletion and inversion efficiency by ddPCR in mature erythroblasts and erythroid progenitors (erythroid burst-forming units [BFU-E]) derived from GFP+ genome-edited healthy donor and SCD HSPCs. Data represent the mean ± SEM of 7 independent experiments. (B) Genotype of single colonies derived from GFP+ HSPCs. The occurrence of deletion and inversion events was assessed in randomly picked BFU-E by PCR (2 healthy donors; n = 100). (C) Sanger sequencing of deletion and inversion junctions in single BFU-E (n = 40). Top rows indicate the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In two alleles, we detected the insertion of 354 bp (*) and 373 bp (^). The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes represent deleted nucleotides. Inserted nucleotides are displayed in lowercase. (D) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from 13.6-kb genome-edited HSPCs. mRNA levels were expressed as fold change vs control cells (ctr-1). γ-Globin expression levels were significantly increased in genome-edited compared with control samples (**P < .01; unpaired Student t test, 2 tailed). A significant β-globin downregulation was detected in edited cells in comparison with control (*P < .05; unpaired Student t test, 2 tailed). δ-Globin expression was decreased in genome-edited samples. Data represent the mean ± SEM of 7 independent experiments. (E) Representative FACS histograms showing the percentage of F-cells and the MFI of HbF immunostaining (in brackets) in mature erythroblasts derived from GFP+ genome-edited (13.6-kb) HSPCs of 2 healthy donors. GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) served as controls. (F) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in genome edited and control samples. The expression of a common AγT chain variant was detected in samples derived from healthy donor 2. The ratio of α chains to non–α chains (in brackets) was unchanged in CRISPR/Cas9-modified samples. (G-H) Quantification of γ- (Aγ+Gγ), β-, and δ- globin protein levels. β-Like globin expression was normalized to α-globin (G). Relative abundance of β-like chains was calculated as percentage of total β-like (β + γ + δ) globins (H). Targeting the 13.6-kb region increased γ-globin chain expression and decreased β-globin protein levels. δ-Globin protein levels were unaffected, suggesting an increased translation of the residual δ-globin transcripts in the absence of β-globin mRNA. Alternatively, the reduced β-chain synthesis favors the incorporation of the δ-globin chain in the Hb tetramers.
Robust HbF upregulation in HSPC-derived erythroblasts upon genome editing of the 13.6-kb region. (A) Assessment of deletion and inversion efficiency by ddPCR in mature erythroblasts and erythroid progenitors (erythroid burst-forming units [BFU-E]) derived from GFP+ genome-edited healthy donor and SCD HSPCs. Data represent the mean ± SEM of 7 independent experiments. (B) Genotype of single colonies derived from GFP+ HSPCs. The occurrence of deletion and inversion events was assessed in randomly picked BFU-E by PCR (2 healthy donors; n = 100). (C) Sanger sequencing of deletion and inversion junctions in single BFU-E (n = 40). Top rows indicate the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In two alleles, we detected the insertion of 354 bp (*) and 373 bp (^). The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes represent deleted nucleotides. Inserted nucleotides are displayed in lowercase. (D) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from 13.6-kb genome-edited HSPCs. mRNA levels were expressed as fold change vs control cells (ctr-1). γ-Globin expression levels were significantly increased in genome-edited compared with control samples (**P < .01; unpaired Student t test, 2 tailed). A significant β-globin downregulation was detected in edited cells in comparison with control (*P < .05; unpaired Student t test, 2 tailed). δ-Globin expression was decreased in genome-edited samples. Data represent the mean ± SEM of 7 independent experiments. (E) Representative FACS histograms showing the percentage of F-cells and the MFI of HbF immunostaining (in brackets) in mature erythroblasts derived from GFP+ genome-edited (13.6-kb) HSPCs of 2 healthy donors. GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) served as controls. (F) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in genome edited and control samples. The expression of a common AγT chain variant was detected in samples derived from healthy donor 2. The ratio of α chains to non–α chains (in brackets) was unchanged in CRISPR/Cas9-modified samples. (G-H) Quantification of γ- (Aγ+Gγ), β-, and δ- globin protein levels. β-Like globin expression was normalized to α-globin (G). Relative abundance of β-like chains was calculated as percentage of total β-like (β + γ + δ) globins (H). Targeting the 13.6-kb region increased γ-globin chain expression and decreased β-globin protein levels. δ-Globin protein levels were unaffected, suggesting an increased translation of the residual δ-globin transcripts in the absence of β-globin mRNA. Alternatively, the reduced β-chain synthesis favors the incorporation of the δ-globin chain in the Hb tetramers.
CRISPR/Cas9-mediated HbF induction ameliorates the SCD cell phenotype
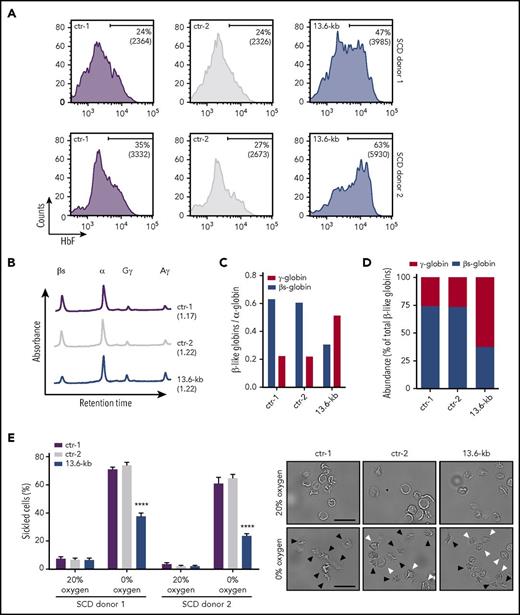
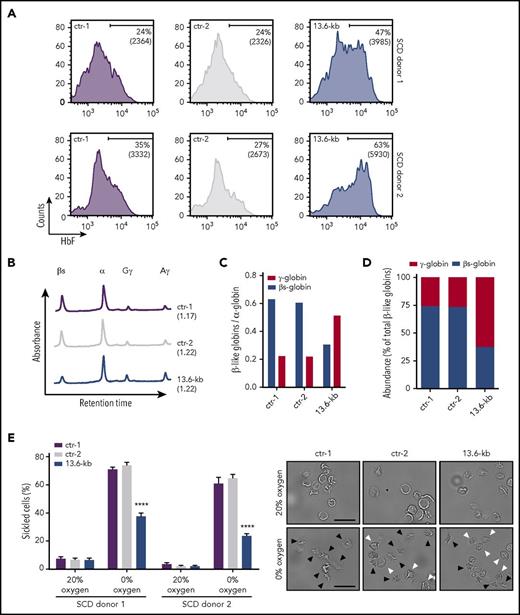
To assess the effect of HbF induction on HbS polymerization, we targeted the 13.6-kb region in bone marrow–derived (SCD donor 1) or mobilized (SCD donor 2) HSPCs from SCD patients. Cells were mobilized by Plerixafor, because administration of G-CSF leads to severe adverse events in SCD patients.44 The deletion and inversion frequency was 17.2% and 15.6% for donor 1 and 34.1% and 28.1% for donor 2. Edited HSPCs were terminally differentiated into enucleated RBCs with no impairment in the erythroid maturation (supplemental Figure 10A-C). FACS analysis showed an increase in the percentage of F-cells and their HbF content in the genome-edited population (Figure 6A). RP-HPLC analysis confirmed a substantial increase of γ-globin chains and a concomitant reduction of βS-globin levels (Figure 6B-C), resulting in an inversion of the β-to-γ globin ratio (Figure 6D). To analyze the effect of HbF upregulation on RBC sickling, we used an in vitro sickling assay that measures the proportion of sickle-shaped RBCs under induced hypoxia. In control SCD cells, induction of HbS polymerization at 0% O2 led to an increase in the fraction of sickled cells up to ∼65% of total RBCs (Figure 6E; supplemental Figure 10D). In the genome-edited RBC population, we observed a lower proportion of sickled cells reaching a maximum of ∼30% of total RBCs (Figure 6E), thus showing that the increase in HbF expression causes an amelioration of the SCD phenotype in cultured cells.
CRISPR/Cas9-mediated induction of HbF expression improves the RBC sickling phenotype. Bone marrow (donor 1) and mobilized (donor 2) SCD CD34+ cells were transfected with plasmids carrying Cas9-GFP and gRNAs targeting the 13.6-kb region. Upon FACS-sorting, GFP+ genome-edited (13.6-kb) HSPCs were terminally differentiated in RBCs using a 3-phase liquid erythroid culture system.24 GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) were used as controls. (A) FACS analysis of HbF expression in control and genome-edited RBCs. The fraction of F-cells and the MFI of HbF immunostaining (in brackets) are displayed in the histograms. (B) RP-HPLC profiles of control and genome-edited RBCs (donor 1). The ratio of α chains to non–α chains is indicated in brackets. (C-D) Quantification of γ (Aγ+Gγ)-globins and sickle β-globin protein levels by RP-HPLC. Globin chain expression was normalized to α-globin (C). Relative abundance of β-like chains was calculated as fraction of total β-like (β + γ) globins (D). (E) In vitro sickling assay measuring the proportion of sickled RBCs under hypoxic conditions (0% O2) (left panel). The percentage of sickled cells was calculated as: (sickled RBC count)/(total RBC count). At least 300 enucleated cells and 10 fields per time point were analyzed for each sample. Data are expressed as mean ± SEM. ****P < .0001 (2-way analysis of variance, Tukey’s multiple comparisons test vs ctr-1 and ctr-2). Representative microscopy images of RBCs before (0 minutes) and after (60 minutes) deoxygenation are shown in the right panel. Black and white arrowheads indicate sickled cells and nonsickled cells, respectively, under hypoxic conditions. Original magnification ×40. Scale bars, 50 µm.
CRISPR/Cas9-mediated induction of HbF expression improves the RBC sickling phenotype. Bone marrow (donor 1) and mobilized (donor 2) SCD CD34+ cells were transfected with plasmids carrying Cas9-GFP and gRNAs targeting the 13.6-kb region. Upon FACS-sorting, GFP+ genome-edited (13.6-kb) HSPCs were terminally differentiated in RBCs using a 3-phase liquid erythroid culture system.24 GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) were used as controls. (A) FACS analysis of HbF expression in control and genome-edited RBCs. The fraction of F-cells and the MFI of HbF immunostaining (in brackets) are displayed in the histograms. (B) RP-HPLC profiles of control and genome-edited RBCs (donor 1). The ratio of α chains to non–α chains is indicated in brackets. (C-D) Quantification of γ (Aγ+Gγ)-globins and sickle β-globin protein levels by RP-HPLC. Globin chain expression was normalized to α-globin (C). Relative abundance of β-like chains was calculated as fraction of total β-like (β + γ) globins (D). (E) In vitro sickling assay measuring the proportion of sickled RBCs under hypoxic conditions (0% O2) (left panel). The percentage of sickled cells was calculated as: (sickled RBC count)/(total RBC count). At least 300 enucleated cells and 10 fields per time point were analyzed for each sample. Data are expressed as mean ± SEM. ****P < .0001 (2-way analysis of variance, Tukey’s multiple comparisons test vs ctr-1 and ctr-2). Representative microscopy images of RBCs before (0 minutes) and after (60 minutes) deoxygenation are shown in the right panel. Black and white arrowheads indicate sickled cells and nonsickled cells, respectively, under hypoxic conditions. Original magnification ×40. Scale bars, 50 µm.
Discussion
Reactivation of HbF by genome editing may be a therapeutic approach to both β-thalassemia and SCD. To identify potential target regions, we carried out a comprehensive analysis of deletional mutations mapped in thalassemic and HPFH individuals and reanalyzed available epigenetic data on histone modifications and transcription factor binding in the β-globin locus. We identified and tested by CRISPR/Cas9-mediated disruption 3 target regions: a 3.5-kb γ-δ intergenic region commonly deleted in HPFH, but not in δβ- and β-thalassemias; the 7.2-kb Corfu deletion leading to elevated HbF levels in β-thalassemic patients; and a more extended 13.6-kb deletion also inactivating the β-globin gene.
The minimal 3.5-kb region binds BCL11A and its partner, GATA1, in adult erythroblasts and it represents a putative HbF silencing element.10 In fetal erythroblasts, GATA1 does not bind the 3.5-kb region, suggesting that it may contribute to the recruitment of BCL11A repressor only in adult erythroblasts. Of note, the evidence that BCL11A occupies this region in primary erythroblasts is conflicting,10,45 leaving the possibility that HbF expression is silenced by other mechanisms. Indeed, the 3.5-kb region contains a 250-bp polypyrimidine-rich sequence targeted by the PYR repressor complex, deletion of which resulted in delayed human γ-to-β-globin switching in some transgenic mouse studies.46-48 Chromatin conformation experiments suggested that the 3.5-kb region establishes the formation of a fetal subdomain and an adult subdomain, thus enabling the LCR to activate globin expression in a stage-specific manner.49 In HPFH, removal of this region might impair the establishment of 2 separate subdomains, thus allowing the LCR to interact also with the γ-globin genes in adult erythroblasts. The 3.5-kb region is contained in the 7.2-kb Corfu deletion, which leads to therapeutically high HbF levels when present in homozygosis in β-thalassemic patients. In heterozygosis, the Corfu deletion in cis to a β-thalassemic mutation causes an approximately threefold elevation of γ-globin primary transcripts that is translated in elevation of γ-globin mRNA and HbF levels only when β-globin mRNA levels fall below a critical threshold.34
Disruption of the 3.5-kb and the Corfu regions by CRISPR/Cas9 editing led to a modest derepression of the fetal γ-globin genes in primary erythroblasts derived from adult HSPCs. The same deletions had no effect in the HUDEP-2 adult erythroblast cell line even when carrying biallelic deletions, suggesting that these cells may not faithfully reproduce all aspects of adult erythropoiesis. Disruption of the putative silencer region containing a potential BCL11A binding site is therefore not sufficient per se to reactivate HbF synthesis at significant levels, suggesting that HbF silencing may be controlled by multiple BCL11A-bound cis-regulatory regions in a redundant fashion.50,51 However, the mild elevation of HbF synthesis in primary erythroblasts was obtained with an editing efficiency of <50% and in cells carrying intact β-globin genes, whereas in patients with increased HbF levels, the Corfu deletion is always associated to a β-thalassemia trait. Further studies are therefore needed to investigate the molecular mechanisms underlying HbF reactivation upon disruption of the 3.5- and 7.2-kb regions.
Disruption of the 13.6-kb region, which extends the Corfu deletion to the first intron of the β-globin gene, caused a robust reactivation of HbF synthesis expression in primary erythroblasts as well as in HUDEP-2 cells. Genome-edited erythroblasts were not counterselected during erythroid differentiation and showed a normal α/non–α-globin chain ratio, indicating that reactivation of HbF compensates for the loss of HbA synthesis and prevents precipitation of uncoupled α-globin chains and ineffective erythropoiesis, consistent with the asymptomatic HPFH phenotype.
Our results indicate that a large deletion spanning the Corfu region, the δ-β intergenic region, and part of the β-globin gene is necessary to reactivate γ-globin gene expression at significant levels. Knockdown of the β-globin gene in primary HSPC-derived erythroblasts produced only a modest effect on HbF expression and caused α/non–α-chain imbalance, as observed in β-thalassemia patients. Deletion of the β-globin promoter in β-thalassemia carriers or of the entire δ-β intergenic region in Senegalese δ0β+-thalassemia and Hb Lepore carriers likewise cause a modest increase in HbF synthesis,52-54 suggesting that β-globin gene inactivation or deletion of the δ-β intergenic region, including the β-globin promoter may represent per se only contributing factors.
A factor contributing to HbF reactivation upon deletion of the 13.6-kb region may be the juxtaposition of the potent 3′ β-globin enhancer to the γ-globin genes. Indeed, 3C experiments in HUDEP-2 clones harboring biallelic deletion of the 13.6-kb region showed an increased interaction of the γ-globin promoter with a region containing the 3′ and intronic β-globin enhancers, suggesting a causal role of these enhancers in activating γ-globin gene expression. However, we cannot exclude an increased interaction of the γ-globin promoter also with the HBBP1 pseudogene and the BGLT3 noncoding RNA gene, both previously described to play a role in HbF regulation50,51 and contained in the fragment assayed in the 3C experiment.
Unexpectedly, inversion of the 13.6-kb region was as effective as its deletion in reactivating HbF synthesis, as analyzed in individual HUDEP-2 clones harboring monoallelic or biallelic inversions and in primary erythroblasts, where almost half of the edited alleles carried an inversion of the 13.6-kb region. Like the deletion, the inversion inactivates the adult β-globin gene, again suggesting that reduced β-globin expression may contribute to a robust increase in HbF synthesis. 3C and histone modification analyses in HUDEP-2 clones harboring either biallelic deletions or inversions showed an increased interaction frequency between LCR and γ-globin promoters, with concomitant increase in H3K27 acetylation. Interestingly, in clones harboring biallelic inversions, the interaction between the LCR and the adult β- and δ-globin gene region was reduced and associated with δ-globin downregulation, indicating that an altered configuration of the 13.6-kb region is associated with re-engagement of the LCR to the fetal γ-gene promoters despite the physical presence of the δ- and β-gene promoters. In addition, inversion of the putative intergenic silencer might impair its repressing activity, as observed for other silencer elements,55,56 and possibly prevent the formation of the fetal and adult subdomains, enabling the LCR to interact with the γ-globin genes. Our study therefore suggests that deletions and inversions may induce chromatin conformation and/or epigenetic changes that ultimately result in γ-globin gene activation.
The effect of locus rearrangement on HbF synthesis was tested in clinically relevant adult HSPCs from healthy donors or SCD patients that were harvested from the bone marrow or mobilized in peripheral blood by G-CSF or Plerixafor administration, respectively. We achieved efficient deletion and inversion of the 13.6-kb region in HSPC-derived erythroblasts, with up to 70% of the BFU-E in clonogenic cultures harboring mono- or biallelic rearrangements. HbF represented up to ∼60% of total hemoglobin in erythroid cells differentiated from genome-edited HSPCs in liquid culture. Edited HSPCs from SCD patients gave rise to functionally corrected mature RBCs in vitro as a result of the concomitant HbF induction and downregulation of HbS synthesis. Overall, these data validate the 13.6-kb region as a potential target to induce a therapeutically-relevant β-to-γ reverse switch for the treatment of β-hemoglobinopathies.
In our study, at least half of the edited cells (∼35% of the total population) carried a biallelic rearrangement as assayed in clonogenic culture, likely resulting in complete correction of the sickling phenotype. The clinical history of allogeneic transplantation in SCD or β-thalassemia patients suggests that a fraction of genetically corrected HSCs of >20% is sufficient to achieve a therapeutic benefit given the in vivo selective survival of corrected RBCs.5,57-59 Whether such a deletion/inversion frequency is achievable in repopulating stem/progenitor cells requires the development of optimized reagents and delivery technology and validation in predictive assays, such as xenotransplantation in appropriate immunodeficient mouse models (ie, mice allowing the engraftment and erythroid differentiation of HSCs60,61 ). Optimization of gRNAs (eg, by the use of alternative protospacers and optimized scaffolds62 ) could be envisioned to increase the proportion of deleted and inverted alleles and minimize off-target effects. Replacement of DNA transfection with less toxic and more efficient delivery of RNA or Cas9 ribonucleoprotein complexes is an additional necessary step toward clinical translation. Of note, we reached up to 45% of deletion/inversion efficiency in unsorted primary HSPC-derived erythroblasts with Cas9 ribonucleoprotein delivery, a level that falls within the window of clinical relevance (A.L., V.M., G.P., Fatima Amor, C.A., T.F., Ciaran Lee, M.P., Gang Bao, M.A., F.M., A.M., manuscript in preparation).
Compared with a classical gene addition approach, such as lentiviral vector-mediated expression of β-globins carrying antisickling mutations,63,64 a genome-editing strategy aimed at forcing a β-to-γ-globin reverse switch would have the advantage of inducing high-level expression of the endogenous γ-globin gene at the expense of the sickle β-globin. Several groups have proposed genome-editing approaches aimed at correcting the SCD mutation,18 disrupting the erythroid-specific BCL11A enhancer14,17 or mimicking genomic deletions or mutations in the γ-globin promoters associated with natural HPFH mutations,13,19,65,66 a strategy validated by clinical genetics. The generation of HPFH-like deletions/inversions described in this study relies on the nonhomologous end joining (NHEJ) repair pathway. This editing strategy might be more efficient as compared with approaches based on homology directed repair18,65-68 or microhomology-mediated end joining,13 given the apparent dominance of the NHEJ repair pathway in HSCs.68,69 Disruption of the erythroid-specific BCL11A enhancer is also based on a NHEJ approach; our study, however, suggests that disruption of the β-globin locus may lead to higher HbF levels,70 although a direct comparison of different studies is difficult and hardly conclusive.
Ultimately, to be validated as therapeutically relevant options for β-hemoglobinopathies, any genome-editing strategy faces a number of potential hurdles in terms of efficacy and safety. These include the development of nontoxic, large-scale editing technology based on clinical-grade reagents and the demonstration of precise editing of a number of long-term repopulating HSCs at least comparable to the efficacious doses predicted by allogeneic transplantation data and currently achievable by classical gene addition technology. Last but not least, an additional advantage of gene editing may potentially be a reduction of the substantial cost currently associated with vector manufacturing in classical gene therapy.
Presented in abstract form at the 58th annual meeting of the American Society of Hematology, San Diego, CA, 4 December, 2016.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jean-Antoine Ribeil for providing the SCD bone marrow samples, Fatima Amor for technical assistance, and Alain Fischer and Isabelle Andre’-Schmutz for discussion and critical reading of the paper.
This work was supported by grants from AFM-Telethon (17224), the European Research Council (ERC-2010-AdG, GT-SKIN, ERC-2015-AdG, GENEFORCURE), Agence Nationale de la Recherché (ANR-16-CE18-0004 and ANR-10-IAHU-01 “Investissements d’avenir” program), and EU Marie Curie-COFUND (PRESTIGE_2015_2_0015) and by a collaboration with CRISPR Therapeutics.
Authorship
Contribution: C.A. and V.M. designed and performed experiments, analyzed data, and wrote the paper; A.L., G.P., T.F., O.R., E.M., L.W., and S.E.H. performed experiments and analyzed data; R.K. and Y.N. provided the HUDEP-2 cell line and protocols for erythroid differentiation; T.J.C., A.S.L., and M.P. contributed to the design of the experimental strategy; M.A., W.E.N., and M.C. performed data analysis and interpretation; F.M. conceived the study and wrote the paper; and A.M. conceived the study, designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: T.J.C. and A.S.L. are employees of and M.P. and F.M. are consultants for CRISPR Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Annarita Miccio, Imagine Institute, 24 Boulevard du Montparnasse, 75015 Paris, France; e-mail: annarita.miccio@institutimagine.org.
References
Author notes
C.A. and V.M. contributed equally to this study.



![Figure 5. Robust HbF upregulation in HSPC-derived erythroblasts upon genome editing of the 13.6-kb region. (A) Assessment of deletion and inversion efficiency by ddPCR in mature erythroblasts and erythroid progenitors (erythroid burst-forming units [BFU-E]) derived from GFP+ genome-edited healthy donor and SCD HSPCs. Data represent the mean ± SEM of 7 independent experiments. (B) Genotype of single colonies derived from GFP+ HSPCs. The occurrence of deletion and inversion events was assessed in randomly picked BFU-E by PCR (2 healthy donors; n = 100). (C) Sanger sequencing of deletion and inversion junctions in single BFU-E (n = 40). Top rows indicate the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In two alleles, we detected the insertion of 354 bp (*) and 373 bp (^). The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes represent deleted nucleotides. Inserted nucleotides are displayed in lowercase. (D) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from 13.6-kb genome-edited HSPCs. mRNA levels were expressed as fold change vs control cells (ctr-1). γ-Globin expression levels were significantly increased in genome-edited compared with control samples (**P < .01; unpaired Student t test, 2 tailed). A significant β-globin downregulation was detected in edited cells in comparison with control (*P < .05; unpaired Student t test, 2 tailed). δ-Globin expression was decreased in genome-edited samples. Data represent the mean ± SEM of 7 independent experiments. (E) Representative FACS histograms showing the percentage of F-cells and the MFI of HbF immunostaining (in brackets) in mature erythroblasts derived from GFP+ genome-edited (13.6-kb) HSPCs of 2 healthy donors. GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) served as controls. (F) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in genome edited and control samples. The expression of a common AγT chain variant was detected in samples derived from healthy donor 2. The ratio of α chains to non–α chains (in brackets) was unchanged in CRISPR/Cas9-modified samples. (G-H) Quantification of γ- (Aγ+Gγ), β-, and δ- globin protein levels. β-Like globin expression was normalized to α-globin (G). Relative abundance of β-like chains was calculated as percentage of total β-like (β + γ + δ) globins (H). Targeting the 13.6-kb region increased γ-globin chain expression and decreased β-globin protein levels. δ-Globin protein levels were unaffected, suggesting an increased translation of the residual δ-globin transcripts in the absence of β-globin mRNA. Alternatively, the reduced β-chain synthesis favors the incorporation of the δ-globin chain in the Hb tetramers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/17/10.1182_blood-2017-10-811505/4/m_blood811505f5.jpeg?Expires=1763578056&Signature=HhdLHOv--ZVZ1jfdZqTBdLj3xHzf3plcQt1L3Ln0wN0XH~MzCHr~j1z81rFsKSLEQvwJkji9gyaunq6rK6msJzVCv9-hCbYCDb9DyNPjvevWZ9wcU4nVVn4L9lBirI5Y62G3e8-KBhc5B~qQemxoyfeN4n4djgSi30WNltGGstrvQw2t~vQE-zpmPXa8SiDvrlumkn9og2CjkcIkuEs72BTOyPyemnuk1kDZOBVvZJDFGCBaMqWLofDtJvuWeHtY7WKhB3NNgIxZ2Qwl7L-D07bxpp8dpoytqZmwKQLMNkJJ6j~N4XeHSF5xRav1F7ESNGogkC3lQC2sM5~NC7v9Yg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. Robust HbF upregulation in HSPC-derived erythroblasts upon genome editing of the 13.6-kb region. (A) Assessment of deletion and inversion efficiency by ddPCR in mature erythroblasts and erythroid progenitors (erythroid burst-forming units [BFU-E]) derived from GFP+ genome-edited healthy donor and SCD HSPCs. Data represent the mean ± SEM of 7 independent experiments. (B) Genotype of single colonies derived from GFP+ HSPCs. The occurrence of deletion and inversion events was assessed in randomly picked BFU-E by PCR (2 healthy donors; n = 100). (C) Sanger sequencing of deletion and inversion junctions in single BFU-E (n = 40). Top rows indicate the predicted junction sequences (in bold). The expected deletion and inversion junctions with small InDels were observed in the majority of genome-edited alleles. In two alleles, we detected the insertion of 354 bp (*) and 373 bp (^). The frequency of each event is calculated as: (number of alleles harboring an identical deletion or inversion junction)/(total number of deleted or inverted alleles). Arrows indicate the predicted junction sites. Dashes represent deleted nucleotides. Inserted nucleotides are displayed in lowercase. (D) qRT-PCR analysis of γ (Aγ+Gγ)-, δ-, and β-globin transcripts in mature erythroblasts derived from 13.6-kb genome-edited HSPCs. mRNA levels were expressed as fold change vs control cells (ctr-1). γ-Globin expression levels were significantly increased in genome-edited compared with control samples (**P < .01; unpaired Student t test, 2 tailed). A significant β-globin downregulation was detected in edited cells in comparison with control (*P < .05; unpaired Student t test, 2 tailed). δ-Globin expression was decreased in genome-edited samples. Data represent the mean ± SEM of 7 independent experiments. (E) Representative FACS histograms showing the percentage of F-cells and the MFI of HbF immunostaining (in brackets) in mature erythroblasts derived from GFP+ genome-edited (13.6-kb) HSPCs of 2 healthy donors. GFP+ cells from Cas9-only samples (ctr-1) and GFP− cells from Cas9+gRNAs cultures (ctr-2) served as controls. (F) RP-HPLC chromatograms showing peaks corresponding to α-globin and β-like globins in genome edited and control samples. The expression of a common AγT chain variant was detected in samples derived from healthy donor 2. The ratio of α chains to non–α chains (in brackets) was unchanged in CRISPR/Cas9-modified samples. (G-H) Quantification of γ- (Aγ+Gγ), β-, and δ- globin protein levels. β-Like globin expression was normalized to α-globin (G). Relative abundance of β-like chains was calculated as percentage of total β-like (β + γ + δ) globins (H). Targeting the 13.6-kb region increased γ-globin chain expression and decreased β-globin protein levels. δ-Globin protein levels were unaffected, suggesting an increased translation of the residual δ-globin transcripts in the absence of β-globin mRNA. Alternatively, the reduced β-chain synthesis favors the incorporation of the δ-globin chain in the Hb tetramers.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/17/10.1182_blood-2017-10-811505/4/m_blood811505f5.jpeg?Expires=1763578057&Signature=mmO5EY13OGRawdK9c~NRx2SxgxPPATDmd3LjFVFT3bQoA4qbWvz17gaLp3RZfKkcMcosF3UqlGouTef-J2rhJkjIrvAbwnYrmS0UNm5ZRyM2LSWZGOL7hqQoOq3ux17momZ3cRhatwJEpCRWr2kI~6qd8fkWoBxiOUg40vOEMIWTbymvQj2dVzPDqFWiJ9QPYOmUnn1gwGh0YJJLLfgIev3E8~pIlsKG40bqO1UxJcQ-TTkvI6WyfvOpEW0eCtySPmM2onO2hyUIiYr0ULwZTTG4kTdJ1bRlNyI~koK8ITom9fUyh9eaYkDF6Kp4qqRmZP9wPu1YJHmzxbBr3ETbOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)