Key Points
Resistance to EZH2 inhibitors occurs due to the activation of survival pathways and acquired EZH2 mutations that prevent drug binding.
Resistance mechanisms for different EZH2 inhibitors varies. Thus, cells resistant to 1 EZH2 inhibitor are sensitive to other inhibitors.
Abstract
Resistance to targeted therapies has become increasingly prevalent. We noted that resistance to different targeted therapies occurs by largely common mechanisms. In this study, we used this information for identifying the mechanisms of resistance to enhancer of zeste homolog 2 (EZH2) inhibitors in diffuse large B-cell lymphoma (DLBCL) harboring EZH2 mutations. We discovered that EZH2 inhibitor-resistant DLBCL cells showed activation of the insulin-like growth factor 1 receptor (IGF-1R), MEK, and phosphoinositide-3-kinase (PI3K) pathways. Constitutive activation of IGF-1R, MEK, or PI3K pathways was sufficient to confer resistance to EZH2 inhibitors in DLBCL. The activation of the PI3K/AKT and MAPK pathways decreased TNFSF10 and BAD expression through a FOXO3-dependent mechanism, which was required for the antitumor effects of EZH2i GSK126. We also identified multiple acquired mutations in EZH2 inhibitor-resistant DLBCL cell lines. These mutations independently conferred resistance to EZH2 inhibitors. Mechanistically, cellular thermal shift assays revealed that the acquired EZH2 mutations that confer resistance to EZH2 inhibitors prevent EZH2 inhibitor binding to the EZH2 mutants. Notably, EZH2 inhibitor GSK126- and EPZ-6438–resistant DLBCL cells remained sensitive to the EZH2 inhibitor UNC1999 and embryonic ectoderm development protein inhibitor EED226, which provides an opportunity to treat DLBCLs that are resistant to these drugs. Collectively, our results underpin the importance for developing a unified approach for forestalling drug resistance by prospectively considering lessons learned from the use of different targeted therapeutic agents.
Introduction
For decades, traditional cytotoxic chemotherapies have been used to treat a wide variety of cancers.1 However, an improved understanding of cancer landscapes has resulted in the identification of cancer cell-specific vulnerabilities, which has led to the development of cancer-specific targeted therapies.2 Nonetheless, despite the benefits provided by targeted therapies, acquired resistance to targeted therapeutic agents, which renders the therapies ineffective, has emerged in a large majority of cancer patients.3 Due to the clinical significance of the problem, tremendous efforts have been devoted to understanding the mechanisms underlying resistance to specific targeted cancer therapies.3,4 Collectively, these studies have identified 2 major mechanisms that drive acquired resistance to targeted therapeutic agents: (1) the activation of prosurvival pathways, and (2) secondary mutations in target proteins that prevent drug binding.5
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of the polycomb-repressive complex 2 (PRC2), which functions as a transcriptional repressor, in part, by methylating histone H3 at lysine 27 (H3K27).6,7 The mutation or overexpression of EZH2 has been identified in several types of cancers and has been shown to correlate with metastatic progression and poor survival.8-12 For example, gain-of-function mutations in EZH2 have been identified in non-Hodgkin lymphomas.11,13 Interestingly, somatic-activating mutations of EZH2 at tyrosine 641 (Y641) within the catalytic SET (Su[var]3-9, Enhancer-of-zeste, and Trithorax) domain alter the substrate specificity, promoting the conversion of H3K27 from dimethylated to trimethylated states, which results in aberrantly high H3K27 trimethylation levels.11,13,14 Small molecule inhibitors of EZH2, such as GSK126 and EPZ-6438 (also known as tazemetostat), have demonstrated strong tumor-suppressive activity against EZH2-mutated lymphomas in preclinical studies.15-17 GSK126 and EPZ-6438 are S-adenosyl-methionine–competitive inhibitors of EZH2 that inhibit the H3K27me3, which consequently leads to the expression of EZH2 target genes.13,15 Similar activities of these EZH2 inhibitors are also observed in other cancer types, such as INI1- and ARID1A-deficient tumors.18
Here, we investigated the role of prosurvival pathways and acquired EZH2 mutations in conferring acquired resistance to EZH2 inhibitors using diffuse large B-cell lymphoma (DLBCL) cells as the model system. We identified several prosurvival pathways and new mutations in EZH2 that confer acquired resistance to EZH2 inhibitors. We also found that DLBCL cells that were resistant to the EZH2 inhibitors GSK126 and EPZ-6438 remained sensitive to the EZH2 inhibitor UNC1999 and embryonic ectoderm development (EED) inhibitor EED226, which suggests a potential treatment option for GSK126- and EPZ-6438–resistant cancer cells. Collectively, our results reveal new opportunities for preventing resistance to EZH2 inhibitors and for treating resistant DLBCL. Furthermore, our results have implications for identifying mechanisms of resistance to new targeted therapies for which the mechanisms underlying resistance remain unclear.
Methods
Generation of EZH2 inhibitor-resistant DLBCL cell lines
DLBCL cell lines (SU-DHL-10, WSU-DLCL-2, and KARPAS-422) were cultured in the presence of the chemical mutagen ethyl methanesulfonate19 (2 mM) for 4 days. The cells were then treated with the EZH2 inhibitor GSK126 (10 µM) for 7 days to enrich the polyclonal GSK-126–resistant DLBCL cells. The cells were then maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin antibiotics. Resistance to EZH2 inhibitors was confirmed by cell viability and apoptosis assay using, respectively, 3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide (MTT) and the apoptosis assay as described in the supplemental Methods (available on the Blood Web site).
Mouse tumorigenesis experiment
KARPAS-422 cells (1 × 107) expressing either EZH2WT, EZH2Y726F, PI3KCA or an empty vector as a control were harvested and resuspended in phosphate-buffered saline (PBS) with 50% Matrigel (BD Biosciences) as previously described by McCabe et al.15 Athymic nude mice (Nu/J; aged 4-6 weeks) were injected subcutaneously with KARPAS-422 cells expressing either EZH2WT, EZH2Y726F, PI3KCA or an empty vector as a control (5 mice per group). Mice were divided into either control (vehicle-treated) or GSK126-treated groups. Vehicle (20% captisol, adjusted to pH 4-4.5 with 1 N acetic acid) or GSK126 dissolved in 20% captisol, adjusted to pH 4-4.5 with 1 N acetic acid was injected intraperitoneally every alternate day (50 mg/kg) for 1 month. Tumor volumes were calculated using the formula: length × width2 × 0.5. All of the procedures related to animal handling, care, and the treatment in this study were approved and performed according to the guidelines of the Institutional Animal Care and Use Committee of Yale University.
Mutational screening of EZH2
Genomic DNA was extracted from both parental and resistant DLBCL cell lines (SU-DHL-10, WSU-DLCL-2, and KARPAS-422) using the DNeasy Blood and Tissue Kit (Qiagen Germantown, MD) according to the manufacturer’s instructions. Polymerase chain reaction amplification of the EZH2 SET domain (from exons 15 to 20) was performed as previously described by Ernst et al using published polymerase chain reaction primers.9 Mutations were identified by sequencing.
Cellular thermal shift assay
A cellular thermal shift assay (CETSA) was conducted as previously described.20-22 Briefly, SU-DHL-10 cells were seeded overnight in RPMI 1640 supplemented with 10% fetal bovine serum and antibiotics. The cells were then treated with incremental concentrations of EZH2 inhibitors for 4 hours. After treatment, the cells were washed in PBS and pelleted by centrifugation at 500g for 2 minutes. The cells were then resuspended in PBS and heated either at 52°C or 37°C for 3 minutes and then cooled at room temperature for 3 minutes as previously described.21 The samples were then lysed and immunoblotting analysis was performed as previously described using the antibodies listed in supplemental Table 2.
Statistical analysis
All of the experiments were conducted in triplicate. The results of experiments are expressed as mean ± standard error of the mean (SEM). An area under the curve (AUC) was calculated to allow for the comparison between 2 curves. The P values were calculated by Student t tests using GraphPad Prism version 6.0h for Macintosh (GraphPad Software, San Diego, CA; www.graphpad.com). Differences were considered when P ≤ .05.
Results
Development of an in vitro model for identifying the mechanisms of acquired resistance to EZH2 inhibitors
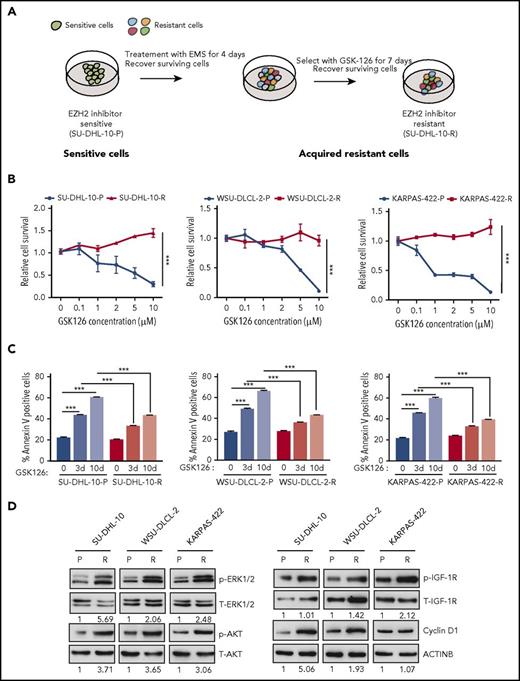
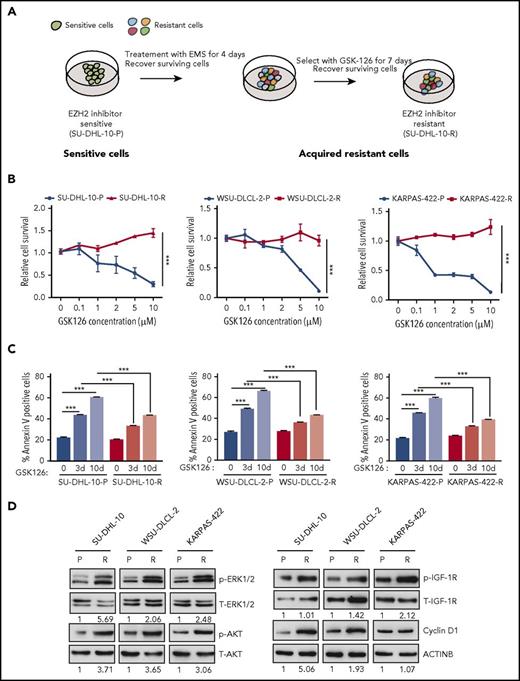
To identify the mechanisms of acquired resistance to EZH2 inhibitors, we developed an in vitro cell culture model. To this end, EZH2 mutant DLBCL cell lines (SU-DHL-10, WSU-DLCL-2, and KARPAS-422) harboring the tyrosine 641 (Y641) mutation in EZH2 were treated with the chemical mutagen ethyl methanesulfonate (EMS)19 for 4 days, followed by treatment with the EZH2 inhibitor GSK126 for 7 days to enrich the resistant cells after mutagenesis (Figure 1A). A proliferation assay revealed that the lymphoma cell lines that were treated first with EMS followed by GSK126 (SU-DHL-10-R, WSU-DLCL-2-R, and KARPAS-422-R) were resistant to GSK126 compared with the parental cell lines (SU-DHL-10-P, WSU-DLCL-2-P, and KARPAS-422-P) (Figure 1B; supplemental Figure 1A). Similarly, GSK126 did not induce apoptosis in GSK126-resistant DLBCL cells (Figure 1C). To show that the GSK126-resistant DLBCL cells were specifically resistant to EZH2 inhibitor, we treated these cells with an unrelated BCL6 inhibitor FX1.23 We found that GSK126-resistant DLBCL cells were sensitive to FX1 (supplemental Figure 1B), which demonstrates that these cells were specifically resistant to EZH2 inhibitor.
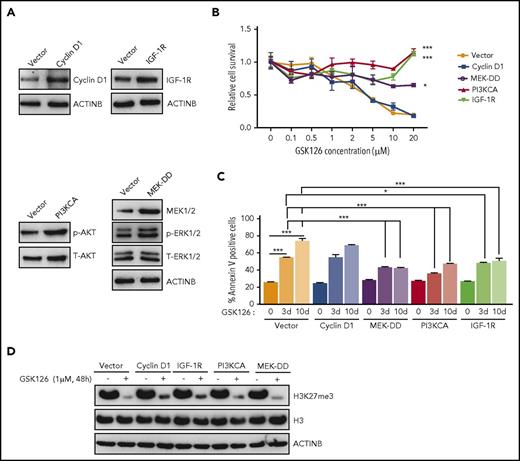
In vitro model of acquired resistance used to identify the mechanisms of resistance to the EZH2 inhibitor GSK126. (A) Schematic for the generation of GSK126-resistant DLBCL cell lines. (B) The parental (P) and GSK126-resistant (R) DLBCL cell lines were treated with the EZH2 inhibitor GSK126 at the indicated concentrations and analyzed for cell viability 72 hours after treatment using an MTT assay. (C) The parental and GSK126-resistant cells were treated with GSK126 (5 μM) for 3 or 10 days and apoptosis was analyzed by annexin V–fluorescein isothiocyanate (FITC) staining. (D) The parental and GSK126-resistant DLBCL cells were analyzed by immunoblotting for the indicated proteins. The densitometric quantitation for phosphorylated ERK1/2 (p-ERK1/2), p-AKT, p-IGF-1R, and cyclin D1 is presented below the respective blots and were normalized to total ERK1/2 (T-ERK1/2), T-AKT, T-IGF-1R, and ACTINB. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. ***P < .001.
In vitro model of acquired resistance used to identify the mechanisms of resistance to the EZH2 inhibitor GSK126. (A) Schematic for the generation of GSK126-resistant DLBCL cell lines. (B) The parental (P) and GSK126-resistant (R) DLBCL cell lines were treated with the EZH2 inhibitor GSK126 at the indicated concentrations and analyzed for cell viability 72 hours after treatment using an MTT assay. (C) The parental and GSK126-resistant cells were treated with GSK126 (5 μM) for 3 or 10 days and apoptosis was analyzed by annexin V–fluorescein isothiocyanate (FITC) staining. (D) The parental and GSK126-resistant DLBCL cells were analyzed by immunoblotting for the indicated proteins. The densitometric quantitation for phosphorylated ERK1/2 (p-ERK1/2), p-AKT, p-IGF-1R, and cyclin D1 is presented below the respective blots and were normalized to total ERK1/2 (T-ERK1/2), T-AKT, T-IGF-1R, and ACTINB. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. ***P < .001.
Several previous studies have suggested important roles of the activated insulin-like growth factor 1 receptor (IGF-1R), MAPK, and phosphoinositide-3-kinase (PI3K) pathways in conferring acquired resistance to targeted cancer therapies.24-26 Additionally, others studies have demonstrated that the overexpression of CCND1 (also known as cyclin D1) can cause resistance to antiestrogen therapies in breast cancer cells and to BRAFV600E inhibitors in melanoma.27,28 To determine whether these mechanisms can also cause resistance to EZH2 inhibitors, we analyzed the PI3K, IGF-1R, and MAPK pathways as well as cyclin D1 expression in parental and GSK126-resistant DLBCL cell lines. We observed that IGF-1R, MAPK, and PI3K signaling were enhanced in GSK126-resistant DLBCL cells compared with parental cells, as observed by increased phosphorylation of IGF-1R, AKT, and extracellular signal-regulated kinase 1/2 (ERK1/2) (Figure 1D). Similarly, we noted increased cyclin D1 expression in GSK126-resistant SU-DHL-10 and WSU-DLCL-2 cells but not in GSK126-resistant KARPAS-422 cells (Figure 1D). Some studies have demonstrated the involvement of autophagy in the development of acquired drug resistance.29,30 Therefore, we did not observe any apparent changes in autophagy-related markers in the GSK126-resistant DLBCL cell lines compared with the parental DLBCL cell lines (supplemental Figure 2A-D). Collectively, these results demonstrated that IGF-1R, MAPK, PI3K signaling, and cyclin D1 expression was enhanced in GSK126-resistant DLBCL cells but autophagy markers remained unchanged.
Activated IGF-1R, PI3K, and MAPK pathways confer resistance to the EZH2 inhibitor
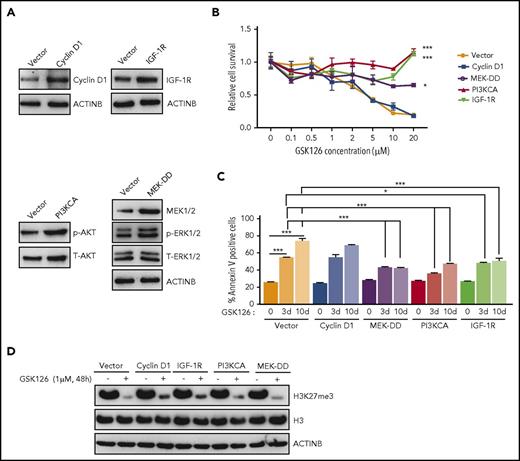
Based on the results presented in Figure 1D, we aimed to determine whether activation of the IGF-1R, PI3K, and MAPK pathways or the increased expression of cyclin D1 could confer resistance to EZH2 inhibitors. Therefore, we ectopically expressed IGF-1R, constitutively active MEK (MEK-DD), a constitutively active form of PI3K (PI3KCA), or cyclin D1 in SU-DHL-10 cells (Figure 2A). After ectopic expression, we measured the cell survival in response to the EZH2 inhibitor GSK126. We found that the ectopic expression of IGF-1R, PI3KCA, or MEK-DD was independently sufficient to confer resistance to GSK126 (Figure 2B; supplemental Figure 2E), whereas the ectopic expression of cyclin D1 failed to do so (Figure 2B; supplemental Figure 2E). Similarly, we also measured the induction of apoptosis in response to GSK126. Our results showed that the cells ectopically expressing IGF-1R, PI3KCA, or MEK-DD exhibited a significantly lower level of apoptosis after GSK126 treatment (Figure 2C).
Activation of the IGF-1R, PI3K, or MAPK pathway is sufficient for conferring resistance to the EZH2 inhibitor GSK126. (A) SU-DHL-10 cells expressing the specified genes or an empty vector were analyzed for the indicated proteins by immunoblotting. (B) SU-DHL-10 cells expressing the specified genes or an empty vector (control) were analyzed for their sensitivity to GSK126 using an MTT assay after treatment with GSK126 at the indicated concentrations for 72 hours. The cell viability relative to DMSO-treated cells is shown. (C) SU-DHL-10 cells expressing the specified genes or an empty vector as a control (Vector) were treated with GSK126 (5 μM) for 3 or 10 days and analyzed for apoptosis induction by annexin V–FITC staining. (D) SU-DHL-10 cells expressing the designated constructs were treated with the EZH2 inhibitor GSK126 (1 μM) for 48 hours and analyzed by immunoblotting for H3K27me3, H3, and ACTINB. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. *P < .05; ***P <.001.
Activation of the IGF-1R, PI3K, or MAPK pathway is sufficient for conferring resistance to the EZH2 inhibitor GSK126. (A) SU-DHL-10 cells expressing the specified genes or an empty vector were analyzed for the indicated proteins by immunoblotting. (B) SU-DHL-10 cells expressing the specified genes or an empty vector (control) were analyzed for their sensitivity to GSK126 using an MTT assay after treatment with GSK126 at the indicated concentrations for 72 hours. The cell viability relative to DMSO-treated cells is shown. (C) SU-DHL-10 cells expressing the specified genes or an empty vector as a control (Vector) were treated with GSK126 (5 μM) for 3 or 10 days and analyzed for apoptosis induction by annexin V–FITC staining. (D) SU-DHL-10 cells expressing the designated constructs were treated with the EZH2 inhibitor GSK126 (1 μM) for 48 hours and analyzed by immunoblotting for H3K27me3, H3, and ACTINB. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. *P < .05; ***P <.001.
Because some studies suggest that restoration of H3K27me3 correlates with resistance to EZH2 inhibitors,31 we also investigated whether IGF-1R, MEK-DD, or PI3KCA overexpression affected H3K27me3 levels in dimethyl sulfoxide (DMSO) or GSK126-treated SU-DHL-10 cells. We found that PI3KCA, MEK-DD, and IGF-1R overexpression was not able to sustain H3K27me3 in the presence of GSK126 despite their ability to induce resistance to EZH2 inhibitors (Figure 2D). These results demonstrated that the activation of IGF-1R, MEK-DD, or PI3K induces resistance to GSK126 through an H3K27me3-independent mechanism in DLBCL.
Determining the outcome of targeting pathways that confer resistance to EZH2 inhibitors in DLBCL cells
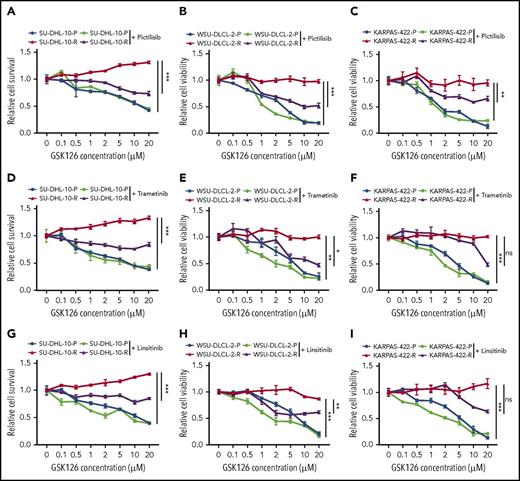
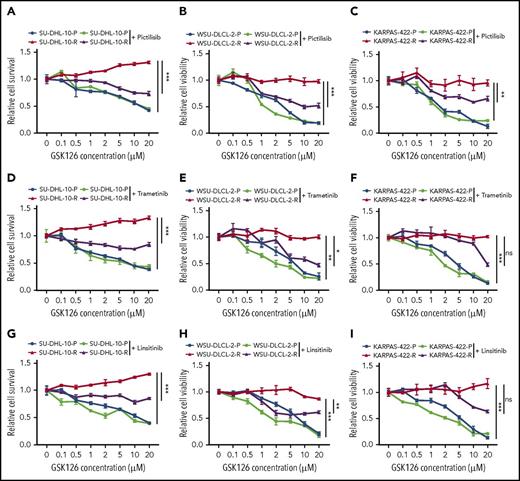
Our studies indicated that the IGF-1R, MAPK, and PI3K pathways could independently confer resistance to the GSK126. Therefore, we wanted to determine whether the individual pharmacological inhibition of these pathways could sensitize the resistant cells to GSK126. To this end, we treated GSK126-resistant DLBCL cell lines with small molecule inhibitors of IGF-1R, MAPK, and PI3K using linsitinib (IGF-1R inhibitor), trametinib (MEK inhibitor), and pictilisib (PI3K inhibitor) in combination with GSK126 in parental and GSK126-resistant DLBCL cell lines. The results of a proliferation assay revealed that GSK126-resistant SU-DHL-10 and WSU-DLCL-2 cell lines were sensitive to GSK126 when treated concurrently with PI3K, IGF-1R, and MEK inhibitors (Figure 3). Despite the beneficial trends of linsitinib and trametinib, only the PI3K inhibitor significantly resensitized the GSK126-resistant KARPAS-422 cell line to GSK126 (Figure 3C). These results show that the cotargeting of some survival pathways, such as the PI3K pathway, in EZH2 inhibitor-resistant DLBCL cells could yield a therapeutic benefit.
Determining the outcome of targeting pathways that confer resistance to EZH2 inhibitors in DLBCL cells. DLBCL cells (SU-DHL-10, WSU-DLCL-2, and KARPAS-422) were treated with GSK126 at the indicated concentrations in combination with 0.1 μM pictilisib (A-C), 0.1 μM trametinib (D-F), or 1 μM linsitinib (G-I) and their sensitivity to GSK126 was determined using an MTT assay 72 hours after treatment. The cell viability relative to DMSO-treated cells is shown. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. *P < .05; **P < .01; ***P < .001. ns, not significant.
Determining the outcome of targeting pathways that confer resistance to EZH2 inhibitors in DLBCL cells. DLBCL cells (SU-DHL-10, WSU-DLCL-2, and KARPAS-422) were treated with GSK126 at the indicated concentrations in combination with 0.1 μM pictilisib (A-C), 0.1 μM trametinib (D-F), or 1 μM linsitinib (G-I) and their sensitivity to GSK126 was determined using an MTT assay 72 hours after treatment. The cell viability relative to DMSO-treated cells is shown. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. *P < .05; **P < .01; ***P < .001. ns, not significant.
EZH2 inhibition induces apoptosis by upregulating the proapoptotic genes TNFSF10 and BAD
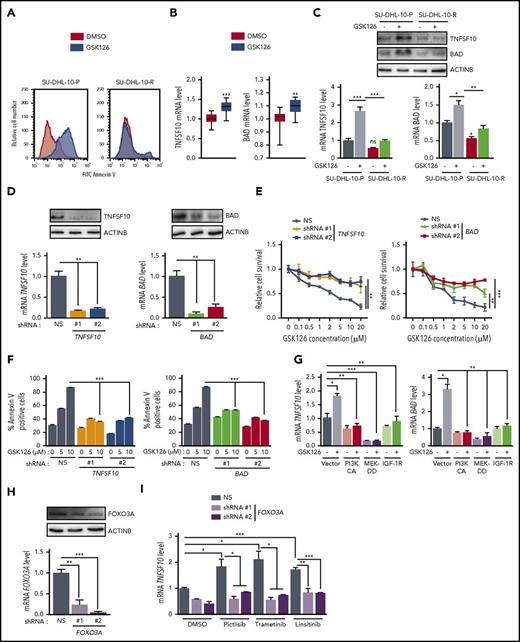
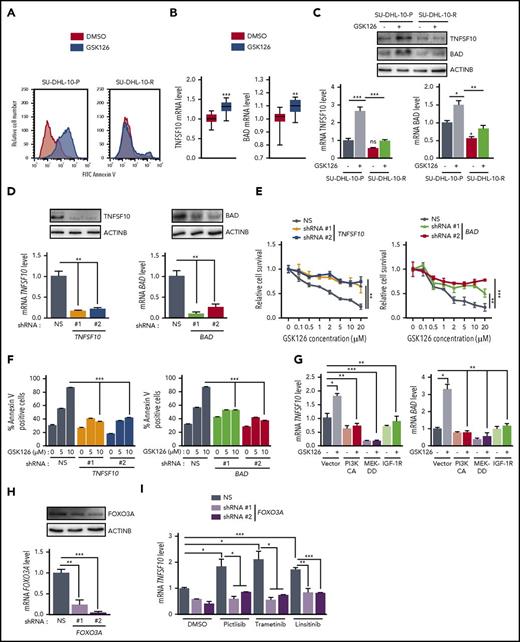
Because EZH2 inhibition decreased cell viability in the parental SU-DHL-10 cell line, we investigated whether this was due to increased apoptosis. Therefore, we measured apoptosis in parental and resistant cell lines after treatment with GSK126. The treatment of parental SU-DHL-10 cells with GSK126 significantly increased the percentage of annexin V+ cells, which suggests an increase in apoptosis, whereas GSK126-resistant SU-DHL-10 cells exhibited a significantly lower percentage of annexin V+ cells in response to GSK126 treatment (Figure 4A).
EZH2 inhibition induces apoptosis by upregulating the proapoptotic genes TNFSF10 and BAD. (A) SU-DHL-10 and SU-DHL-10-P cells were treated with GSK126 (10 μM) for 3 days and apoptosis was analyzed by annexin V–FITC staining. (B) TNFSF10 and BAD mRNA expression in previously published Gene Expression Omnibus (GEO) datasets including DLBCL cells treated with GSK126 (GSE: 40971). (C) TNFSF10 and BAD mRNA and protein expressions in SU-DHL-10-P and SU-DHL-10-R cells treated with GSK126 (1 μM, 48 hours). (D) TNFSF10 and BAD mRNA (bottom) or protein (top) expression in SU-DHL-10 cells expressing nonsilencing (NS) or the designated shRNAs. (E) SU-DHL-10 cells expressing nonsilencing or TNFSF10 and BAD shRNAs were analyzed using an MTT assay after treatment with the indicated concentrations of GSK126 for 72 hours. Relative proliferation under the specified conditions is shown. (F) SU-DHL-10 cells expressing the designated shRNAs were treated with vehicle or GSK126 (5 and 10 μM) for 72 hours and apoptosis was measured using an annexin V assay. (G) TNFSF10 and BAD mRNA expression in SU-DHL-10 cells expressing either PI3KCA, MEK-DD, or IGF-1R under the specified conditions. (H) FOXO3A mRNA (bottom) and protein (top) expression in SU-DHL-10 cells expressing nonsilencing or FOXO3A shRNAs. (I) TNFSF10 mRNA expression in SU-DHL-10 cells expressing nonsilencing or FOXO3A shRNAs treated with the indicated drugs for 48 hours. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001.
EZH2 inhibition induces apoptosis by upregulating the proapoptotic genes TNFSF10 and BAD. (A) SU-DHL-10 and SU-DHL-10-P cells were treated with GSK126 (10 μM) for 3 days and apoptosis was analyzed by annexin V–FITC staining. (B) TNFSF10 and BAD mRNA expression in previously published Gene Expression Omnibus (GEO) datasets including DLBCL cells treated with GSK126 (GSE: 40971). (C) TNFSF10 and BAD mRNA and protein expressions in SU-DHL-10-P and SU-DHL-10-R cells treated with GSK126 (1 μM, 48 hours). (D) TNFSF10 and BAD mRNA (bottom) or protein (top) expression in SU-DHL-10 cells expressing nonsilencing (NS) or the designated shRNAs. (E) SU-DHL-10 cells expressing nonsilencing or TNFSF10 and BAD shRNAs were analyzed using an MTT assay after treatment with the indicated concentrations of GSK126 for 72 hours. Relative proliferation under the specified conditions is shown. (F) SU-DHL-10 cells expressing the designated shRNAs were treated with vehicle or GSK126 (5 and 10 μM) for 72 hours and apoptosis was measured using an annexin V assay. (G) TNFSF10 and BAD mRNA expression in SU-DHL-10 cells expressing either PI3KCA, MEK-DD, or IGF-1R under the specified conditions. (H) FOXO3A mRNA (bottom) and protein (top) expression in SU-DHL-10 cells expressing nonsilencing or FOXO3A shRNAs. (I) TNFSF10 mRNA expression in SU-DHL-10 cells expressing nonsilencing or FOXO3A shRNAs treated with the indicated drugs for 48 hours. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001.
To understand the mechanism of GSK126-mediated apoptosis induction, we analyzed previously published data that evaluated gene expression patterns in EZH2 mutant DLBCL cells after GSK126 treatment.15 The ingenuity pathway analysis of altered genes from this study revealed the modulation of several apoptotic genes, including the tumor necrosis factor/tumor necrosis factor receptor superfamily genes TNFSF10 and BCL-2–associated agonist of cell death (BAD) (Figure 4B; supplemental Table 1; supplemental Figure 3).
We first analyzed TNFSF10 and BAD messenger RNA (mRNA) and protein levels after treating parental and GSK126-resistant SU-DHL-10 cell lines with GSK126. Our results confirmed that EZH2 inhibition increases TNFSF10 and BAD expression in parental SU-DHL-10 cells (Figure 4C). Interestingly, GSK126 failed to induce the increased expression of the proapoptotic TNFSF10 and BAD in GSK126-resistant cells (Figure 4C).
Next, we tested the ability of EZH2 inhibitors to induce apoptosis through TNFSF10 and/or BAD in DLBCL cells. To this end, we knocked down the expression of TNFSF10 and BAD via short hairpin RNAs (shRNAs) in SU-DHL-10 cells (Figure 4D). We then performed a proliferation assay after treating SU-DHL-10 cells expressing TNFSF10 or BAD shRNAs with GSK126 and found that the loss of either TNFSF10 or BAD conferred resistance to GSK126 compared with SU-DHL-10 cells expressing a nonspecific shRNA (Figure 4E). Based on these results, we also measured the induction of apoptosis. We found that the induction of apoptosis by GSK126 was significantly inhibited after TNFSF10 or BAD knockdown (Figure 4F). These results demonstrate that GSK126 requires TNFSF10 and BAD to induce apoptosis and that both are important mediators of the antitumor effects of GSK126.
We then investigated the regulation of TNFSF10 and BAD expression in EZH2 inhibitor-resistant cells. Because we found that IGF-1R, PI3K, and MAPK confer resistance to GSK126 through an EZH2-independent mechanism, we sought to determine the role of these pathways in the regulation of TNFSF10 and BAD expression. We individually overexpressed IGF-1R, PI3KCA, and MEK-DD in SU-DHL-10 cells and treated the cells with GSK126 for 48 hours and analyzed TNFSF10 and BAD mRNA levels. Our results indicated that the activation of the IGF-1R, PI3K, and MAPK pathways hindered the induction of TNFSF10 and BAD expression by GSK126 (Figure 4G).
Previous studies have shown that the inhibition of the PI3K/AKT and MAPK/ERK pathways induces the expression of proapoptotic genes, such as TNFSF10, by promoting the nuclear translocation of the transcription factor FOXO3A.32 Therefore, we tested whether the IGF-1R, MAPK, and PI3K pathways regulate TNFSF10 expression through a FOXO3A-dependent pathway. To test this possibility, we knocked down FOXO3A expression using shRNAs in SU-DHL-10 cells and treated them with either pictilisib, trametinib, or linsitinib. Our results showed that the knockdown of FOXO3A impaired the ability of the PI3K, MEK, and IGF-1R inhibitors to induce TNFSF10 expression (Figure 4H-I). Our results confirmed that these pathways regulate TNFSF10 through FOXO3A. We also analyzed the phosphorylation of FOXO3A at Ser253 by immunoblotting in parental and resistant SU-DHL-10 cells after GSK126 treatment and observed increased FOXO3A phosphorylation in the GSK126-resistant SU-DHL-10 cells (supplemental Figure 4). Collectively, these results demonstrate that the activation of survival pathways in GSK126-resistant cells inhibits TNFSF10 via a FOXO3A-dependent mechanism and impairs the ability of GSK126 to induce apoptosis in resistant cells due to IGF-1R, PI3K, and MAPK activation.
Acquired mutations in EZH2 prevent drug binding
The analysis of SU-DHL-10-P and SU-DHL-10-R cells revealed that EZH2 expression was higher in GSK126-resistant cells (SU-DHL-10-R) and that GSK126 treatment did not effectively inhibit H3K27me3 (Figure 5A). Additionally, GSK126 treatment failed to rescue the expression of EZH2 target genes in SU-DHL-10-R cells following GSK126 treatment (Figure 5B). These effects could possibly be due to acquired mutations in EZH2 that render GSK126 ineffective and stabilize EZH2. Secondary mutations have been shown to play a critical role in conferring resistance to targeted therapies.33,34
Role of EZH2 mutations in conferring resistance to the EZH2 inhibitor. (A) SU-DHL-10-P or SU-DHL-10-R cells were treated with either DMSO or with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 expression. Histone H3 and ACTINB were used as loading controls. (B) The mRNA expression of the indicated EZH2 target genes in SU-DHL-10-P or SU-DHL-10-R cells that were treated with either DMSO or with GSK126 (1 μM) for 48 hours. (C) Gene and protein schematics for EZH2 and the mutations identified from the genomic DNA sequencing of the S-adenosyl-methionine domain are shown. (D) DNA-sequencing chromatograms reveal the presence of the C663Y, E720G, and Y726F mutations in EZH2 in GSK126-resistant DLBCL cell lines. (E) Table of previously reported EZH2 mutations found in sequenced cancer samples. Data were collected from the cBioPortal for the designated studies. (F) Mapping of the EZH2C663Y, EZH2E720G, and EZH2Y726F mutations on to the structure of the EZH2 SET domain (Protein Data Bank [PDB] ID: 4MI542 ) are shown. (G) A CETSA was performed for the EZH2WT, EZH2Y641F, EZH2C663Y, EZH2E720G, and EZH2Y726F mutants in SU-DHL-10 cells. The samples were analyzed for EZH2 and ACTINB by immunoblotting. The densitometric quantitation for EZH2 at 52°C is presented below the respective blots and were normalized to ACTINB. The P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001. AA, amino acid; ID, identification; Mut, mutation; NA, not available.
Role of EZH2 mutations in conferring resistance to the EZH2 inhibitor. (A) SU-DHL-10-P or SU-DHL-10-R cells were treated with either DMSO or with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 expression. Histone H3 and ACTINB were used as loading controls. (B) The mRNA expression of the indicated EZH2 target genes in SU-DHL-10-P or SU-DHL-10-R cells that were treated with either DMSO or with GSK126 (1 μM) for 48 hours. (C) Gene and protein schematics for EZH2 and the mutations identified from the genomic DNA sequencing of the S-adenosyl-methionine domain are shown. (D) DNA-sequencing chromatograms reveal the presence of the C663Y, E720G, and Y726F mutations in EZH2 in GSK126-resistant DLBCL cell lines. (E) Table of previously reported EZH2 mutations found in sequenced cancer samples. Data were collected from the cBioPortal for the designated studies. (F) Mapping of the EZH2C663Y, EZH2E720G, and EZH2Y726F mutations on to the structure of the EZH2 SET domain (Protein Data Bank [PDB] ID: 4MI542 ) are shown. (G) A CETSA was performed for the EZH2WT, EZH2Y641F, EZH2C663Y, EZH2E720G, and EZH2Y726F mutants in SU-DHL-10 cells. The samples were analyzed for EZH2 and ACTINB by immunoblotting. The densitometric quantitation for EZH2 at 52°C is presented below the respective blots and were normalized to ACTINB. The P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001. AA, amino acid; ID, identification; Mut, mutation; NA, not available.
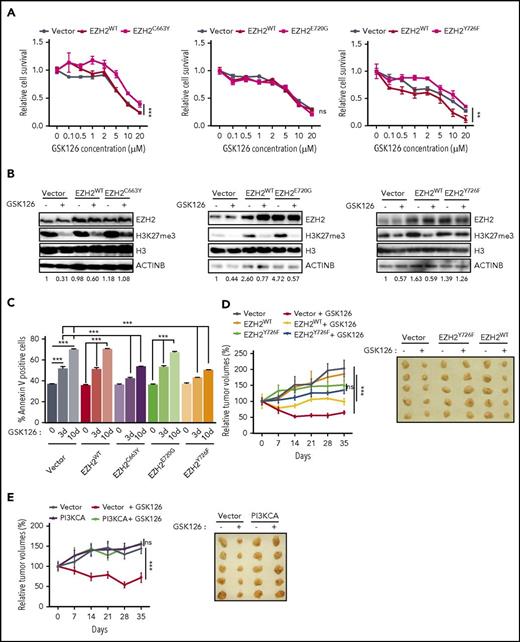
To test this possibility, we sequenced the EZH2 SET domain using genomic DNA isolated from parental and GSK126-resistant DLBCL cell lines. After identifying acquired mutations in the SET domain (Figure 5C-D; supplemental Figure 5), we focused on 3 specific EZH2 mutations because they were repeated in multiple GSK126-resistant DLBCL cell lines, such as the single TAC→TTC substitution of tyrosine for phenylalanine (EZH2Y726F) at codon 726, or they were identified by The Cancer Genome Sequencing (TCGA) in some cancer types (Figure 5E). The latter included the TGC→TAC substitution of cysteine for tyrosine (EZH2C663Y) at codon 663 and the GAA→GGA substitution of glutamic acid for glycine (EZH2E720G) at codon 720 (Figure 5C-E; supplemental Figure 5). We then performed structural analysis to assess the consequences of these 3 mutations. The structural analysis indicated that the EZH2C663Y and EZH2Y726F mutations might alter the binding of the EZH2 inhibitor to EZH2 and that EZH2E720G might not have an effect on EZH2 inhibitor binding (Figure 5F).
Based on these results, we tested the direct effects of the acquired mutations on EZH2 inhibitor binding. To this end, we generated EZH2C663Y, EZH2E720G, and EZH2Y726F mutants by site-directed mutagenesis and stably expressed EZH2 wild-type (EZH2WT) or each of the 3 mutants individually in SU-DHL-10 cells. To determine whether these mutants prevented drug binding, we performed a CETSA, which is an established technique for determining the impact of mutations on drug binding.20,35 Based on previous studies,21,22 we used 52°C as the temperature for determining whether GSK126 induced dose-dependent thermal stabilization of EZH2WT, EZH2Y641F, EZH2E720G, EZH2C663Y, and EZH2Y726F. Our results demonstrated that GSK126 binds to EZH2WT, EZH2Y641F, and EZH2E720G (Figure 5G). However, EZH2C663Y and EZH2Y726F inhibited the drug-induced thermal stabilization of EZH2, which indicates that GSK126 was not able to bind to these mutants (Figure 5G). Collectively, our data demonstrated that the acquisition of the EZH2C663Y and EZH2Y726F mutations is sufficient to inhibit the binding of GSK126 to EZH2.
Acquired EZH2 mutations confer resistance to EZH2 inhibitors
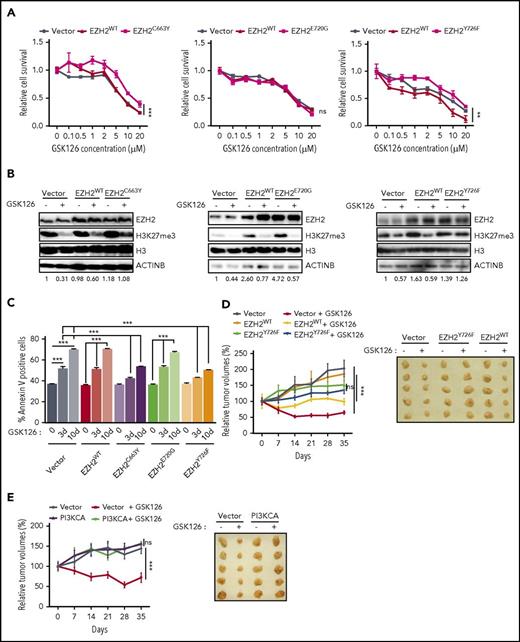
Next, we tested whether EZH2C663Y and EZH2Y726F affected the ability of GSK126 and EPZ-6438 to inhibit the growth of DLBCL cells expressing these mutants. Our results showed that both EZH2C663Y and EZH2Y726F conferred resistance to EZH2 inhibitors, whereas neither EZH2E720G nor EZH2WT expression affected the ability of either GSK126 or EPZ-6438 to inhibit proliferation (Figure 6A; supplemental Figures 6A and 7A). We also investigated the ability of each mutant to maintain the H3K27me3 mark in the presence of either GSK126 or EPZ-6438. We found that cells expressing the EZH2C663Y and EZH2Y726F mutants were able to sustain H3K27me3 even in the presence of EZH2 inhibitors (GSK126 and EPZ-6438), whereas EZH2E720G failed to maintain H3K27 methylation (Figure 6B; supplemental Figure 7B). Consistent with our earlier results, we also observed significantly lower apoptosis in DLBCL cells expressing the EZH2C663Y and EZH2Y726F mutants after GSK126 or EPZ-6438 treatment whereas neither EZH2E720G nor EZH2WT affected the ability of GSK126 or EPZ-6438 to induce apoptosis (Figure 6C; supplemental Figure 7C). Because the EZH2 mutations at C663 and Y726 yield a higher level of the H3K27me3 modification, we measured the mRNA expression of TNFSF10 and BAD in cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, or EZH2Y726F in the presence and absence of GSK126. We found that the cells overexpressing either EZH2C663Y or EZH2Y726F failed to induce the expression of TNFSF10 or BAD in the presence of GSK126 (supplemental Figure 6B-C). Only cells stably expressing EZH2WT or EZH2E720G significantly induced the expression of proapoptotic genes after GSK126 treatment (supplemental Figure 6B-C).
EZH2 mutations prevent EZH2 inhibitor binding. (A) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or empty vector were treated with the specified concentrations of GSK126 for 72 hours and assayed for cell viability using an MTT assay. The cell viability compared with untreated cells is shown. (B) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or an empty vector were treated with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 by immunoblotting. Histone H3 and ACTINB were used as loading controls. The densitometric quantitation for H3K27me3 is presented below the respective blots and were normalized to total histone H3. (C) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or an empty vector were treated with GSK126 (5 μM) for 3 or 10 days and apoptosis was analyzed by annexin V–FITC staining. (D-E) Athymic nude mice were subcutaneously injected with KARPAS-422 cells expressing either empty vector, EZH2WT, EZH2Y726F (D) or with empty vector or PI3KCA (E) and treated with GSK126 (50 mg/kg) every alternate day for 31 days. Mean tumor volume ± SEM (n = 5) and representative tumor images for the experiments presented in panels D and E under the indicated conditions are shown. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. **P < .01; ***P < .001.
EZH2 mutations prevent EZH2 inhibitor binding. (A) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or empty vector were treated with the specified concentrations of GSK126 for 72 hours and assayed for cell viability using an MTT assay. The cell viability compared with untreated cells is shown. (B) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or an empty vector were treated with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 by immunoblotting. Histone H3 and ACTINB were used as loading controls. The densitometric quantitation for H3K27me3 is presented below the respective blots and were normalized to total histone H3. (C) SU-DHL-10 cells overexpressing either EZH2WT, EZH2C663Y, EZH2E720G, EZH2Y726F, or an empty vector were treated with GSK126 (5 μM) for 3 or 10 days and apoptosis was analyzed by annexin V–FITC staining. (D-E) Athymic nude mice were subcutaneously injected with KARPAS-422 cells expressing either empty vector, EZH2WT, EZH2Y726F (D) or with empty vector or PI3KCA (E) and treated with GSK126 (50 mg/kg) every alternate day for 31 days. Mean tumor volume ± SEM (n = 5) and representative tumor images for the experiments presented in panels D and E under the indicated conditions are shown. AUC was calculated to allow for the comparison between the 2 curves and the P values were calculated using a Student t test. Data are presented as mean ± SEM. **P < .01; ***P < .001.
To characterize the EZH2C663Y or EZH2Y726F mutations further, we tested whether the GSK126-resistant SU-DHL-10 cells were resistant to UNC1999, another EZH2 inhibitor that does not interact with the amino acid residue 663.36 The results of the proliferation assays revealed that both SU-DHL-10-R and SU-DHL-10 cells ectopically expressing EZH2C663Y were sensitive to UNC1999-mediated growth inhibition (supplemental Figure 8A-B). However, SU-DHL-10 cells overexpressing EZH2Y726F were resistant to UNC1999 treatment (supplemental Figure 8C). Similarly, only the EZH2Y726F mutation was able to significantly block apoptosis in response to GSK126 whereas neither SU-DHL-10-R nor SU-DHL-10 cells ectopically expressing EZH2WT, EZH2C663Y, EZH2E720G affect the ability of UNC1999 to induce apoptosis (supplemental Figure 8D-E). The results of the immunoblotting analysis showed that cells overexpressing EZH2C663Y were not able to maintain H3K27me3 levels after UNC1999 treatment compared with cells expressing EZH2Y726F (supplemental Figure 8F-G). In accord with these results, the CETSA confirmed that UNC1999 binds to EZH2C663Y but not to EZH2Y726F (supplemental Figure 9). These results show that in some cases, DLBCL cells that are resistant to 1 EZH2 inhibitor remain sensitive to other EZH2 inhibitors, which could mean that switching the treatment regimen from 1 inhibitor to another might overcome drug resistance in some cases. Recently, EED226, a newly developed allosteric inhibitor of the PRC2 complex protein EED, has been shown to inhibit cells that are resistant to EZH2 inhibitors.37 Therefore, we also tested this new compound for its ability to inhibit EZH2 inhibitor-resistant cells. Interestingly, proliferation and apoptosis assays showed that GSK126-resistant DLBCL cell lines remained sensitive to EED226 (supplemental Figure 10).
Finally, we validated our key findings using a mouse model of DLBCL cell xenogaft. To this end, we subcutaneously injected KARPAS-422 cells expressing either empty vector, EZH2WT, EZH2Y726F or with empty vector or PI3KCA into the flank of athymic nude mice. These mice were then treated with GSK126. Our results showed that although GSK126 significantly inhibited the growth of tumor xenograft expressing an empty vector or EZH2WT (Figure 6D-E), tumor xenografts of DLBCL cells expressing EZH2Y726F mutant or PI3KCA conferred resistance to GSK126 treatment (Figure 6D-E). Collectively, these results further confirm our findings in cell culture.
Discussion
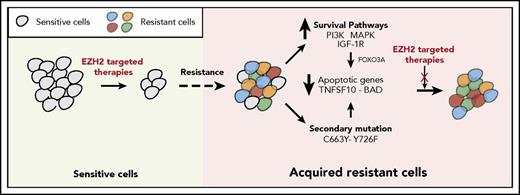
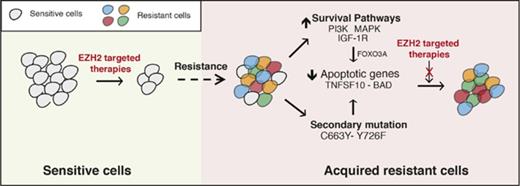
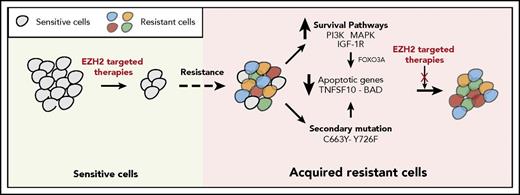
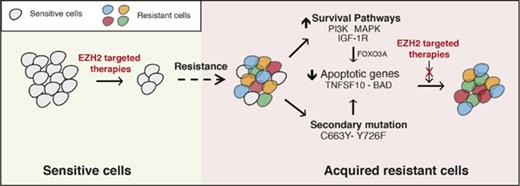
The failure of targeted therapeutic agents due to the emergence of drug resistance is a significant clinical problem. In this study, we uncovered the mechanisms underlying resistance to EZH2 inhibitors using previously identified mechanisms of resistance to other targeted therapeutic agents as a guide. We found that the mechanisms of resistance to targeted therapeutic agents, including EZH2 inhibitors, fall into 2 major categories that include the activation of prosurvival pathways (IGF-1R, PI3K, and MAPK) and the emergence of EZH2 mutations that prevent drug binding (Figure 7).
Mechanisms of resistance to EZH2 inhibitors. The model shows the mechanism by which IGF-1R, PI3K and MAPK, or EZH2 mutations confer resistance to EZH2 inhibitors in DLBCL cells.
Mechanisms of resistance to EZH2 inhibitors. The model shows the mechanism by which IGF-1R, PI3K and MAPK, or EZH2 mutations confer resistance to EZH2 inhibitors in DLBCL cells.
Several small molecule inhibitors that directly target either EZH2 or other components of the PRC2 complex, such as EED, have been developed.37,38 Most EZH2 inhibitors, including GSK126, EPZ-6438, and UNC1999, are S-adenosyl methionine competitive inhibitors that block EZH2 methyltransferase activity.16,39 We found that the activation of the IGF-1R, PI3K, and MEK pathways was sufficient to cause resistance to EZH2 inhibitors. Additionally, we discovered mutations in the EZH2 gene, some of which rendered DLBCL cells resistant to GSK126 and EPZ-6438 by preventing drug binding. Furthermore, we found that acquired resistance to EZH2 inhibitors works, in part, by inhibiting the proapoptotic proteins TNFSF10 and BAD via a FOXO3A-dependent mechanism. Additionally, we and others have found that both TNFSF10 and BAD are regulated by EZH2 and prosurvival pathway targets.40,41
Our studies show that targeting single resistance mechanisms such as the IGF-1R or PI3K pathways is beneficial. Additionally, we found that most of the survival pathways that we identified as well as the EZH2 acquired mutations that conferred resistance to EZH2 inhibitor all hindered the activation of BAD and TNFSF10. Therefore, activating these proapoptotic pathways might be of significantly higher clinical value compared with targeting either a single prosurvival pathway or an EZH2 mutation associated with acquired resistance to EZH2 inhibitors.
Interestingly, we found that EZH2 mutations that arose in GSK126-resistant cells also rendered the cells resistant to both GSK126 and EPZ-6438 and prevented their binding. However, SU-DHL-10 cells expressing some of these mutations, including EZH2C663Y or EZH2E720G, were still sensitive to UNC1999. These results indicate that there are differences between EZH2 inhibitors regarding the mechanisms by which they cause acquired drug resistance. This provides a potentially useful clinical opportunity that can be exploited to treat tumors that acquire resistance to 1 EZH2 inhibitor but might still be sensitive to another inhibitor. To this end, EED226, a newly developed allosteric inhibitor of the PRC2 complex protein EED, has been shown to inhibit cells that are resistant to EZH2 inhibitors.37 We also found that DLBCL cells ectopically expressing EZH2WT, EZH2C663Y, EZH2E720G, or EZH2Y726F remained sensitive to EED226.
Collectively, our studies provide new opportunities to identify mechanisms of drug resistance for newly developed targeted therapeutics for which the mechanisms of resistance are still not known. Based on the results of our study, we recommend that the lessons learned regarding the mechanisms of resistance identified following the previous use of different targeted therapeutic agents should be factored prospectively in drug design and the administration of drug combinations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Titus Boggon for the structural analysis presented in Figure 5 and supplemental Figure 5. The authors thank Parmanand Malvi and Suresh Bugide for help with animal experiments.
This work was supported by National Institutes of Health (NIH), National Cancer Institute grants R01CA195077-01A1 (N.W.), R01CA200919-01 (N.W.), and an NIH supplement. N.W. was also supported by American Cancer Society Research Scholar grant 128347-RSG-15-212-01-TBG. This work was also supported by the Elsa U. Pardee Foundation, a Development Research Project from the Yale Specialized Programs of Research Excellence in Lung Cancer, and NIH National Cancer Institute administrative supplement 3P50CA196530-02S1.
Authorship
Contribution: M.B. and N.W. conceived and designed the experiments, analyzed and interpreted the data, and cowrote the manuscript; and M.B. performed all of the experiments, prepared the figures, and performed the statistical analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Narendra Wajapeyee, Department of Pathology, School of Medicine, Yale University, 310 Cedar St, New Haven, CT, 06510; e-mail: narendra.wajapeyee@yale.edu.

![Figure 5. Role of EZH2 mutations in conferring resistance to the EZH2 inhibitor. (A) SU-DHL-10-P or SU-DHL-10-R cells were treated with either DMSO or with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 expression. Histone H3 and ACTINB were used as loading controls. (B) The mRNA expression of the indicated EZH2 target genes in SU-DHL-10-P or SU-DHL-10-R cells that were treated with either DMSO or with GSK126 (1 μM) for 48 hours. (C) Gene and protein schematics for EZH2 and the mutations identified from the genomic DNA sequencing of the S-adenosyl-methionine domain are shown. (D) DNA-sequencing chromatograms reveal the presence of the C663Y, E720G, and Y726F mutations in EZH2 in GSK126-resistant DLBCL cell lines. (E) Table of previously reported EZH2 mutations found in sequenced cancer samples. Data were collected from the cBioPortal for the designated studies. (F) Mapping of the EZH2C663Y, EZH2E720G, and EZH2Y726F mutations on to the structure of the EZH2 SET domain (Protein Data Bank [PDB] ID: 4MI542) are shown. (G) A CETSA was performed for the EZH2WT, EZH2Y641F, EZH2C663Y, EZH2E720G, and EZH2Y726F mutants in SU-DHL-10 cells. The samples were analyzed for EZH2 and ACTINB by immunoblotting. The densitometric quantitation for EZH2 at 52°C is presented below the respective blots and were normalized to ACTINB. The P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001. AA, amino acid; ID, identification; Mut, mutation; NA, not available.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/19/10.1182_blood-2017-08-804344/4/m_blood804344f5.jpeg?Expires=1769098332&Signature=LbyWsGed7LwpdmeEVpmqTC3pnNsjyv0V49HFzG34ok9BeMTxO0YH5hSklcATd7s0Rx7MBbKCKdV~zneCtLB7yvKCkg9wCjc-CnzQxNuiJVHnu7HU1gKVccctgfonCEDAlhTgaXGaVnx2Idcl6SwNd1qteJZ2D7t-dp7jRmUpyyRmSf5fwDw6VIzD8lPDGUZIvVdtd4VpKOPQ0FeafsoEb7figjUsGX0ALeRu3lFgl2NJcktXHUpDOM3ZzhxXegaqHwHV~4dpf0Gswvjyi50cejoIbAcm4YAF7NvILkBgTMhL-fCaIQz2Gy28dEczTK2xx9aw0H-g5~dzoWpByIVLrA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. Role of EZH2 mutations in conferring resistance to the EZH2 inhibitor. (A) SU-DHL-10-P or SU-DHL-10-R cells were treated with either DMSO or with GSK126 (1 μM) for 48 hours and analyzed for EZH2 and H3K27me3 expression. Histone H3 and ACTINB were used as loading controls. (B) The mRNA expression of the indicated EZH2 target genes in SU-DHL-10-P or SU-DHL-10-R cells that were treated with either DMSO or with GSK126 (1 μM) for 48 hours. (C) Gene and protein schematics for EZH2 and the mutations identified from the genomic DNA sequencing of the S-adenosyl-methionine domain are shown. (D) DNA-sequencing chromatograms reveal the presence of the C663Y, E720G, and Y726F mutations in EZH2 in GSK126-resistant DLBCL cell lines. (E) Table of previously reported EZH2 mutations found in sequenced cancer samples. Data were collected from the cBioPortal for the designated studies. (F) Mapping of the EZH2C663Y, EZH2E720G, and EZH2Y726F mutations on to the structure of the EZH2 SET domain (Protein Data Bank [PDB] ID: 4MI542) are shown. (G) A CETSA was performed for the EZH2WT, EZH2Y641F, EZH2C663Y, EZH2E720G, and EZH2Y726F mutants in SU-DHL-10 cells. The samples were analyzed for EZH2 and ACTINB by immunoblotting. The densitometric quantitation for EZH2 at 52°C is presented below the respective blots and were normalized to ACTINB. The P values were calculated using a Student t test. Data are presented as mean ± SEM; *P < .05; **P < .01; ***P < .001. AA, amino acid; ID, identification; Mut, mutation; NA, not available.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/19/10.1182_blood-2017-08-804344/4/m_blood804344f5.jpeg?Expires=1769176631&Signature=TNwFGFl3iFVbWSTT10vn9p4OraGt~w9winVN7~kIjJrODhSamcG9DIsP154Xtk4oimQWoaroEw3jP0UJ3~BL-xWJ8fBFT3NywAhMm4Zv8FCjodTxGnxSIT~-TIzCYE3RduUaCPFEEDVnWeNYC7J98NGJRAfkwpTL-Q3EOxPlTMiJWIBL0qfz0J3nW-86P44XUvxEnvlwrmUN34tPCiBmbhRoX7ATMJQcjhx~tKXaXSsrEd9agB3psSeo4IcfZON~L5evmqPkmyN1hSNMyEQRq6941cBcqb03y7CaCezO33DsTAf9-~TfYf-0dQ9D-pwDSM~UucAjZyqhTLR9NqvTXw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)