Key Points
ctDNA is as an easily accessible source of tumor DNA for cHL genotyping.
ctDNA is a radiation-free tool to track residual disease in cHL.
Abstract
The rarity of neoplastic cells in the biopsy imposes major technical hurdles that have so far limited genomic studies in classical Hodgkin lymphoma (cHL). By using a highly sensitive and robust deep next-generation sequencing approach for circulating tumor DNA (ctDNA), we aimed to identify the genetics of cHL in different clinical phases, as well as its modifications on treatment. The analysis was based on specimens collected from 80 newly diagnosed and 32 refractory patients with cHL, including longitudinal samples collected under ABVD (adriamycin, bleomycin, vinblastine, dacarbazine) chemotherapy and longitudinal samples from relapsing patients treated with chemotherapy and immunotherapy. ctDNA mirrored Hodgkin and Reed-Sternberg cell genetics, thus establishing ctDNA as an easily accessible source of tumor DNA for cHL genotyping. By identifying STAT6 as the most frequently mutated gene in ∼40% of cases, we refined the current knowledge of cHL genetics. Longitudinal ctDNA profiling identified treatment-dependent patterns of clonal evolution in patients relapsing after chemotherapy and patients maintained in partial remission under immunotherapy. By measuring ctDNA changes during therapy, we propose ctDNA as a radiation-free tool to track residual disease that may integrate positron emission tomography imaging for the early identification of chemorefractory patients with cHL. Collectively, our results provide the proof of concept that ctDNA may serve as a novel precision medicine biomarker in cHL.
Introduction
An unprecedented body of genetic knowledge has been translated into biomarkers to refine diagnosis, prognostication, and treatment of non-Hodgkin lymphomas (NHLs).1 On the contrary, the genetics of classical Hodgkin lymphoma (cHL) is less well understood.2 Rarity of neoplastic Hodgkin and Reed-Sternberg (HRS) cells in the biopsies and their routine formalin fixation impose major technical hurdles that have limited the assessments of cHL mutations in different clinical phases and under different treatments.
An unmet medical need in cHL is the early and accurate identification of chemorefractory patients, as they are candidates for treatment intensification to maximize the chances of cure, as well as the early and accurate identification of good-risk patients, as they are candidates for treatment de-escalation to avoid long-term complications of chemoradiotherapy.3
Interim positron emission tomography/computed tomography (PET/CT) is widely used to predict cHL outcome before completion of chemotherapy and to inform early treatment intensification or de-escalation. However, interim PET/CT results are inconsistent with the final outcome in ∼20% to 30% of patients, who are thus still exposed to over- or undertreatment.4 By identifying residual disease beyond the sensitivity of imaging and by specifically tracking tumor fingerprints, molecular methods hold the potential of complementing PET/CT in assessing tumor response.5 However, molecular minimal residual disease has been so far inapplicable in lymphomas that lack a leukemic component, such as cHL.
Plasma is a source of circulating tumor DNA (ctDNA) for genotyping purposes,6 and studies using copy number abnormalities,7 immunoglobulin gene rearrangement,8 or single mutations as tumor marker9 identified ctDNA also in cHL. After having shown that ctDNA mirrors HRS cells’ mutational profile, we expanded the applications of ctDNA in cHL by showing that it can be used to noninvasively detect cHL mutations without the need for HRS cell microdissection, longitudinally track cHL clonal evolution under different treatments, and monitor residual disease during multiagent chemotherapy with ABVD (adriamycin, bleomycin, vinblastine, dacarbazine).
Methods
Patients
The study had a retrospective observational design. Confirmed diagnosis of cHL and availability of biological samples were the sole study inclusion criteria. cHL subtyping was according to the World Health Organization Classification of Tumors of Hematopoietic and Lymphoid Tissues criteria.10 Both previously untreated (n = 80) and relapsed/refractory (n = 32) patients with cHL were included. All relapsed/refractory cHL failed both first-line therapy as well as autologous transplant salvage. The following biological material was analyzed: cell-free DNA (cfDNA) isolated from plasma collected at diagnosis before treatment start (n = 80 patients), during ABVD courses (on day 1 of course 2 at the time of interim reassessment, and at the end of treatment; n = 24 patients), at refractory progression (n = 32 patients), before and after failing autotransplant salvage (n = 6), before and after failing brentuximab vedotin (n = 5 patients), and before and during therapy with nivolumab (n = 5 patients); and normal germline genomic DNA (gDNA) from peripheral blood granulocytes collected at the same time of the paired plasma sample. For comparative purposes, tumor gDNA from HRS cells and gDNA from biopsy areas devoid of HRS cells were also analyzed. Paired plasma samples were collected in close temporal proximity of the tumor tissue biopsy (7-14 days after the biopsy). cfDNA from newly diagnosed diffuse large B-cell lymphoma (DLBCL; n = 19) and primary mediastinal large B-cell lymphoma (n = 3) and the corresponding paired normal gDNA were also analyzed. DLBCLs have been previously published and now reanalyzed with a different target region.11 Patients provided informed consent in accordance with local Institutional Review Board requirements and the Declaration of Helsinki. The study was approved by the Cantonal Ethical Committee of Ticino (CE 3236, BASEC 2017-01004).
Microdissection, immunohistochemistry, and in situ hybridization
The percentage of tumor cells was visually estimated independently by 2 pathologists (L.M.L. and M. Martini) on hematoxylin and eosin-stained slides. For each case, multiple specific tumor areas containing 20% to 40% HRS cells were microdissected to enrich the proportion of neoplastic cells, and were pooled. Marking the tumor enriched-regions within tissue samples relied on the visual assessment of tissue sections or CD30 immunohistochemical staining of HRS cells. Areas devoid of HRS cells were also microdissected for comparative purposes. Expression of pSTAT6 and MCH-I was assessed by immunohistochemistry and scored as previously reported (supplemental Appendix, available on the Blood Web site).12,13 In situ hybridization of Epstein-Barr virus–encoded small RNAs (EBERs) on formalin-fixed and paraffin-embedded tissue section was carried out as described.14
CAPP-seq library preparation, ultra-deep NGS, and variant calling
To avoid preanalytic confounding effects, a standardized approach was used to extract cfDNA from plasma (supplemental Appendix). A targeted resequencing gene panel including coding exons and splice sites of 77 genes (target region: 191 271 bp) that are recurrently mutated in mature B-cell tumors has been specifically designed for this project (supplemental Table 1) as follows: initial seed genes were chosen if recurrently mutated in more than 5% of mature B-cell tumors, in the case of well-known cancer genes (even if mutated in 1%-5% of cases), or if they were known to be associated with resistance to chemotherapy in mature B-cell tumors; and for genes with a well-defined hotspot, only coding exons plus splice sites that included more than 95% of mutations were covered. Tumor and germline gDNA from tissues (median = 450 ng) were sheared through sonication before library construction to obtain 200-bp fragments. For cfDNA, which is a naturally fragmented DNA, a median amount of 79.63 ng was used for library construction without additional fragmentation. The next-generation sequencing (NGS) libraries were constructed using the KAPA Library Preparation Kit (Kapa Biosystems), and hybrid selection was performed with the custom SeqCap EZ Choice Library (Roche NimbleGen). In the CAncer Personalized Profiling by deep Sequencing (CAPP-seq) of cfDNA, the manufacturer’s protocols were modified as previously reported.15 Multiplexed libraries were sequenced using 150- or 300-bp paired-end runs on NextSeq or MiSeq sequencers (Illumina). A robust and previously validated bioinformatics pipeline was used for variant calling (supplemental Appendix).11
Validation of hotspot STAT6 mutation
Hotspot STAT6 mutations (c.1249A>T and c.1255G>A) discovered by CAPP-seq in cfDNA were validated by allele-specific polymerase chain reaction (supplemental Appendix).
Imaging studies
PET/CT scans were performed at baseline for initial staging, after 2 cycles of ABVD and at the end of treatment, or at baseline, after 4, 10, 17, and 25 cycles of nivolumab. PET and CT images were acquired in the same session. CT scans obtained with a low-dose protocol were used for attenuation-correction of the PET images. All patients were fasting for at least 6 hours before the injection of 250 to 370 MBq (4.5 MBq/kg) 18FDG (fluorodeoxyglucose). Blood glucose measured before injection of the radiotracer was lower than 160 mg/dL in all patients. PET data were acquired in 2- or 3-dimensional mode from the midthigh toward the base of the skull after a standardized uptake time of 60 minutes (±5 minutes). The PET acquisition time was at least 3 minutes per bed position. Images were reconstructed with validated and commercially available iterative algorithms, and standardized uptake values were automatically calculated. A central blind analysis was performed by trained nuclear physicians (L.C., S.A., V.R., and A.G.) without information on the clinical outcome. The 18FDG-PET/CT images obtained at baseline and during treatment were analyzed following a standard protocol on a dedicated workstation (Siemens Syngo MMWP Workstation VE36A; Siemens). The interim and end-of-treatment and 18FDG-PET/CT images scans were visually assessed according to the Deauville criteria, with 18FDG uptake of any residual lesion scored according to the 5-point scale, using mediastinal blood pool and liver uptake as reference settings.16 The LYmphoma Response to Immunomodulatory therapy Criteria (LYRIC) were applied on treatment with nivolumab.17 Diffuse uptake in the spleen or marrow on the postchemotherapy scan that was considered a result of chemotherapy was not scored as active disease.
Statistical analysis
Progression-free survival was measured from date of ABVD treatment start to date of progression (event), death from any cause (event), or last follow-up (censoring). Overall survival was measured from date of ABVD start to date of death (event) or last follow-up (censoring). Molecular studies were blinded to the study endpoints. Survival analysis was performed by Kaplan-Meier method. Unsupervised hierarchical clustering (Euclidean distance, complete method) according to the mutational status was performed using the extended package in the R environment (https://cran.r-project.org/package=dendextend; R Studio console; RStudio, Boston, MA). Categorical variables were compared by χ-square test and Fisher’s exact test. Continuous variables were compared by Mann-Whitney U test. All statistical tests were 2-sided. Statistical significance was defined as P < .05. Correction for multiple comparisons was performed by using the false-discovery-rate method. The analysis was performed with SPSS v.22 and with R statistical package 3.3.2 (http://www.r-project.org).
Results
Circulating tumor DNA mirrors the genetics of HRS cells
The study was based on specimens collected from 80 newly diagnosed patients and 32 patients with relapsed-refractory cHL, including longitudinal samples obtained under ABVD chemotherapy and from relapsing patients treated with chemotherapy and immunotherapy. We retrospectively profiled a total of 349 blood and tissue samples
To provide the proof that ctDNA informs on cHL genetics, it was initially genotyped using CAPP-seq, a sensitive (∼10−3) and validated targeted ultra-deep-NGS approach for ctDNA,11,15 in a discovery set of 15 patients (mean coverage: 4941×; supplemental Figure 1A) provided with paired tumor gDNA isolated from HRS cells. Paired normal gDNA was also analyzed to confirm the somatic origin of mutations (mean coverage: 4626×; supplemental Figure 1B). High concordance (R2 = 0.978) of variant calling from independent duplicate experiments supported the robustness of CAPP-seq of ctDNA, including detection of low-abundance mutations (supplemental Figure 1D), thus excluding their origin from a batch-specific experimental noise.
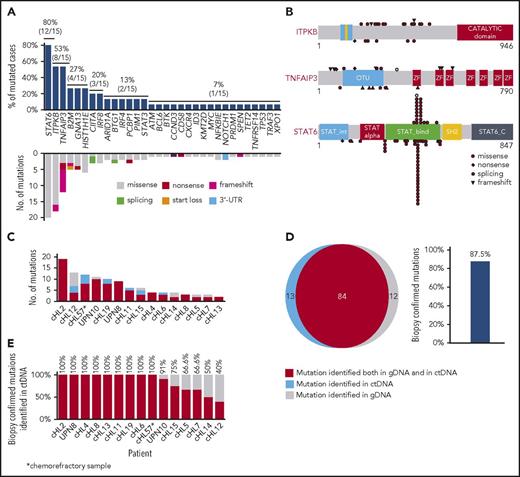
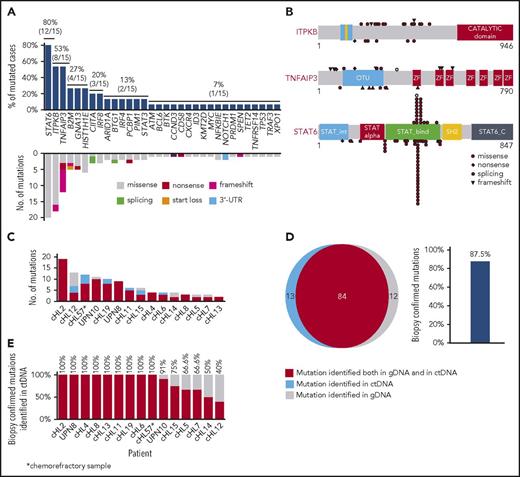
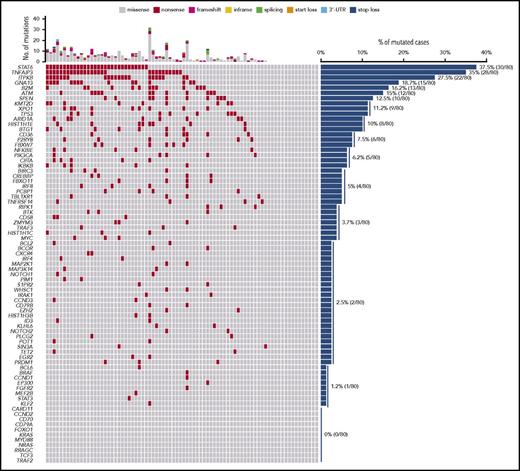
Nonsynonymous somatic mutations were discovered in ctDNA of all 15 patients with cHL, including variants of the TNFAIP3, ITPKB, GNA13, and B2M genes, which were previously reported in studies of purified HRS cells (Figure 1A).18,19 The molecular spectrum of mutations discovered in ctDNA also reflected that of published cHL variants (Figure 1A-B), thus supporting their tumor origin.18,19
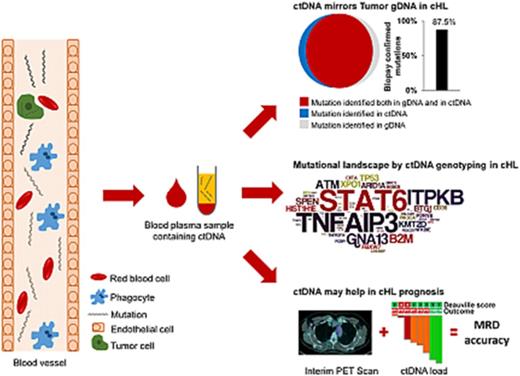
ctDNA mirrors the genetics of cHL. (A) Prevalence of nonsynonymous somatic mutations discovered in ctDNA of 15 cHL cases provided with HRS cells microdissected from the paired biopsy. The graph below shows the number and type of nonsynonymous somatic mutations identified in each gene. (B) The position and type of nonsynonymous somatic mutations identified by ctDNA genotyping of the most frequently mutated genes are reported at the top of the proteins. The position and type of nonsynonymous somatic mutations that have been detected in the tumor gDNA of published cHL series19 (ITPKB and TNFAIP3) or B-cell lymphomas of the COSMIC database (version 81; STAT6) are reported at the bottom of the protein. Shapes indicate the type of the mutations, whereas color codes indicate whether they have been identified in the paired microdissected HRS cells (red) or they lacked in the paired microdissected HRS cells (gray). (C) Number of mutations in a given tumor discovered in plasma ctDNA and/or tumor gDNA. Mutations are color coded if they were identified in both plasma ctDNA and tumor gDNA (red), only in plasma ctDNA (blue), or only in tumor gDNA (gray). (D) Venn diagram summarizing the overall number of mutations discovered in both plasma ctDNA and tumor gDNA (red), only in plasma ctDNA (blue), or only in tumor gDNA (gray). The corresponding overall sensitivity of plasma cfDNA genotyping in discovering biopsy-confirmed mutations is shown. (E) For each patient, the fraction of tumor biopsy-confirmed mutations that were detected in plasma ctDNA is shown. Patients are ordered by decreasing detection rates. The red portion of the bars marks the prevalence of tumor biopsy-confirmed mutations that were detected in plasma ctDNA. The gray portion of the bars marks the prevalence of tumor biopsy-confirmed mutations that were not detected in plasma ctDNA.
ctDNA mirrors the genetics of cHL. (A) Prevalence of nonsynonymous somatic mutations discovered in ctDNA of 15 cHL cases provided with HRS cells microdissected from the paired biopsy. The graph below shows the number and type of nonsynonymous somatic mutations identified in each gene. (B) The position and type of nonsynonymous somatic mutations identified by ctDNA genotyping of the most frequently mutated genes are reported at the top of the proteins. The position and type of nonsynonymous somatic mutations that have been detected in the tumor gDNA of published cHL series19 (ITPKB and TNFAIP3) or B-cell lymphomas of the COSMIC database (version 81; STAT6) are reported at the bottom of the protein. Shapes indicate the type of the mutations, whereas color codes indicate whether they have been identified in the paired microdissected HRS cells (red) or they lacked in the paired microdissected HRS cells (gray). (C) Number of mutations in a given tumor discovered in plasma ctDNA and/or tumor gDNA. Mutations are color coded if they were identified in both plasma ctDNA and tumor gDNA (red), only in plasma ctDNA (blue), or only in tumor gDNA (gray). (D) Venn diagram summarizing the overall number of mutations discovered in both plasma ctDNA and tumor gDNA (red), only in plasma ctDNA (blue), or only in tumor gDNA (gray). The corresponding overall sensitivity of plasma cfDNA genotyping in discovering biopsy-confirmed mutations is shown. (E) For each patient, the fraction of tumor biopsy-confirmed mutations that were detected in plasma ctDNA is shown. Patients are ordered by decreasing detection rates. The red portion of the bars marks the prevalence of tumor biopsy-confirmed mutations that were detected in plasma ctDNA. The gray portion of the bars marks the prevalence of tumor biopsy-confirmed mutations that were not detected in plasma ctDNA.
To validate our approach and formally confirm the tumor origin of ctDNA variants, HRS cell-enriched areas (tumor representation, 20%-40%) were microdissected from the paired tumor biopsies and their gDNA analyzed by CAPP-seq blinded to the mutational profile recovered in the paired ctDNA. To systematically derive the accuracy of ctDNA genotyping, the results of plasma ctDNA genotyping and tumor gDNA genotyping (gold standard) were then compared. Mean coverage of the tumor gDNA was 1733× (supplemental Figure 1C). Genotyping of plasma ctDNA identified a total of 106 somatic mutations, whereas genotyping of the gDNA from HRS cells identified 96 somatic mutations (Figure 1C-D). No mutations were detected in areas of the biopsy devoid of HRS cells, thus validating mutation origin from HRS cells. Biopsy-confirmed tumor mutations were detectable in ctDNA samples with an 87.5% (n = 84/96; 95% confidence interval, 79.2%-92.8%) sensitivity (Figure 1D-E). A few variants were not identified in tumor gDNA. These variants were bona fide neoplastic, according to their molecular profile (Figure 1B) and disappearance on disease remission. Their absence in microdissected HRS cells was conceivably a result of the subclonal or anatomical heterogeneity of the tumor, as already extensively reported in other ctDNA studies.11,15,20
Noninvasive genotyping of newly diagnosed cHL
After having established that ctDNA mirrors tumor gDNA in cHL, we used this source of tumor DNA to noninvasively characterize the mutational spectrum of 80 newly diagnosed patients whose clinicopathologic characteristics are summarized in Table 1. We detected nonsynonymous somatic mutations in 81.2% of patients, with an average of 5 mutations per case. The lack of correlation between number of mutations and coverage or input cfDNA for library preparation (supplemental Figure 2A-B) indicated that the conditions of our CAPP-seq reached a plateau in sensitivity that was uniformly maintained across all the study samples. The mean allele frequency of ctDNA mutations was 5.5% (range, 0.29%-74.0%), and the majority (87.3%) had an allele frequency higher than 1% (supplemental Figure 1C). Pretreatment ctDNA concentration correlated with a stage and prognostic group of cHL, thus pointing to ctDNA as a surrogate marker of tumor load (supplemental Figure 3).
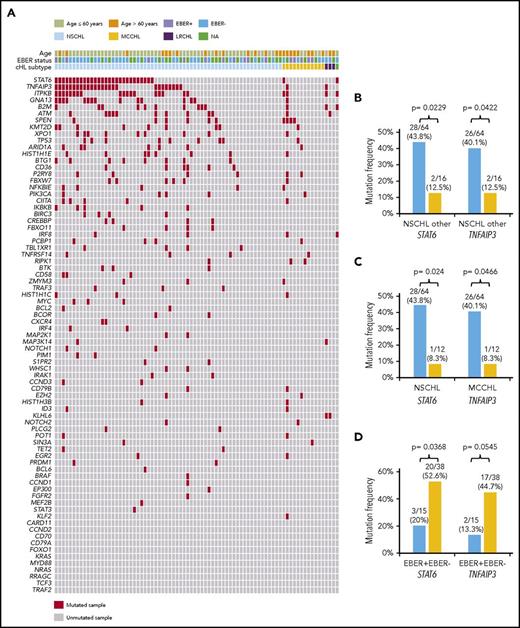
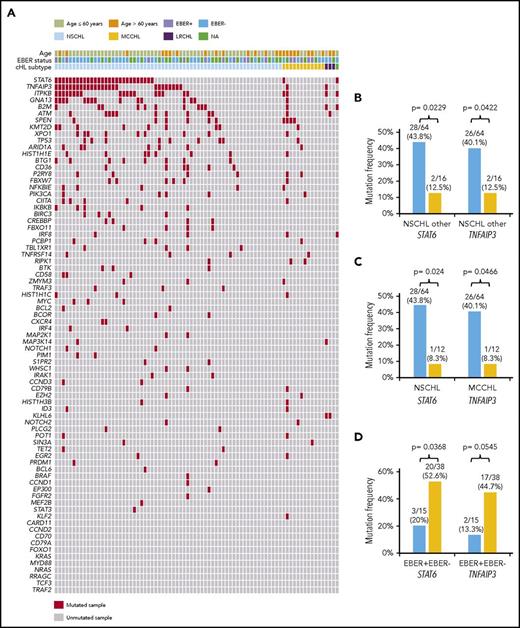
Genes recurrently affected by nonsynonymous somatic mutations in 20% or more of patients included STAT6 (37.5%), TNFAIP3 (35.0%), and ITPKB (27.5%; Figure 2; supplemental Table 2), which often co-occurred in the same patient (Figure 2; supplemental Figure 4). Hotspot STAT6 mutations (c.1249A>T, n = 26; c.1255G>A, n = 12) discovered in cfDNA were confirmed on a different experimental platform by allele-specific polymerase chain reaction, thus validating CAPP-seq results (supplemental Figure 5). STAT6 and TNFAIP3 mutations were enriched in nodular sclerosis cHL compared with other cHL subtypes, including mixed cellularity cases, and among EBER-negative cHL compared with EBER-positive cases (Figure 3), further reinforcing the notion that cHL subtypes have different genetic profiles.18,19 STAT6 mutations also were more frequent among patients aged 60 years or younger, which might reflect the enrichment of mixed cellularity and EBER-positive cases among elderly cHL (supplemental Figure 6).
The mutational profile of newly diagnosed cHL. The heat map shows individual nonsynonymous somatic mutations detected in ctDNA of newly diagnosed cHL (n = 80). Each row represents a gene, and each column represents a primary tumor. The heat map was manually clustered to emphasize mutational co-occurrence. Mutations are color-coded in red. The number and type of nonsynonymous somatic mutations in any given patients is plotted above the heat map. The horizontal bar graph shows the gene mutation frequency.
The mutational profile of newly diagnosed cHL. The heat map shows individual nonsynonymous somatic mutations detected in ctDNA of newly diagnosed cHL (n = 80). Each row represents a gene, and each column represents a primary tumor. The heat map was manually clustered to emphasize mutational co-occurrence. Mutations are color-coded in red. The number and type of nonsynonymous somatic mutations in any given patients is plotted above the heat map. The horizontal bar graph shows the gene mutation frequency.
Distribution of mutations among cHL histologic subtypes. (A) The heat map shows individual nonsynonymous somatic mutations detected in ctDNA of newly diagnosed cHL (n = 80) clustered according to the histological subtype. Each row represents a gene, and each column represents a primary tumor. The heat map was manually clustered to emphasize mutational co-occurrence. Mutations are color-coded in red. EBER status is shown above the heat map. The bar graph compares the prevalence of the most frequently mutated genes between nodular sclerosis (NSCHL) and all the other cHL subtypes (B), between nodular sclerosis (NSCHL) and mixed cellularity (MCCHL) cHL (C), and between EBER positive and negative cases (D).
Distribution of mutations among cHL histologic subtypes. (A) The heat map shows individual nonsynonymous somatic mutations detected in ctDNA of newly diagnosed cHL (n = 80) clustered according to the histological subtype. Each row represents a gene, and each column represents a primary tumor. The heat map was manually clustered to emphasize mutational co-occurrence. Mutations are color-coded in red. EBER status is shown above the heat map. The bar graph compares the prevalence of the most frequently mutated genes between nodular sclerosis (NSCHL) and all the other cHL subtypes (B), between nodular sclerosis (NSCHL) and mixed cellularity (MCCHL) cHL (C), and between EBER positive and negative cases (D).
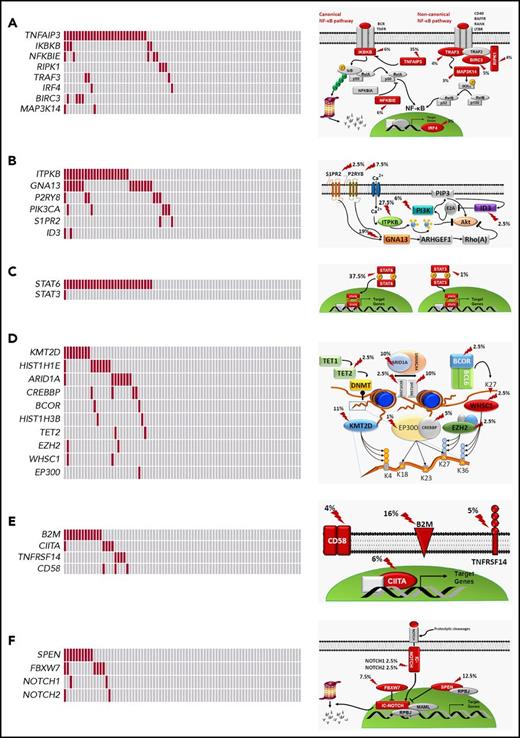
Mutated genes pointed to the molecular deregulation of specific programs in cHL. NF-κB was mutated in 46.2% of patients, consistent with the strong NF-κB signature of HRS cells (Figure 4A).21,22 At variance with DLBCL, cHL was virtually devoid of mutations in upstream molecules that converge surface signals to NF-κB and that sensitize tumor cells to Bruton tyrosine kinase inhibition (Figure 2).23 PI3K/AKT was affected in 46.2% of patients, consistent with the preclinical evidence that cHL is addicted to this actionable cellular program (Figure 4B).24,25 Cytokine signaling was mutated in 37.5% of patients (Figure 4C), although the prevalence of genetic lesions affecting this pathway may be higher and not fully captured by our target region that did not cover the SOCS1 gene. A number of epigenetic genes were cumulatively affected in 35.0% of patients, with the exception of EZH2, whose druggable mutations were rare in cHL (Figures 2 and 4D).26 Genes involved in immune surveillance were mutated in 27.5% of patients (Figure 4E). The NOTCH pathway was mutated in 20.0% of patients (Figure 4F), in keeping with the NOTCH signature of HRS cells.25,27
Pathways that are recurrently mutated in newly diagnosed cHL. In the heat map, rows correspond to genes and columns represent individual patients. Color coding is based on gene alteration status (gray, wild type; red, mutated). The heat map was manually clustered to emphasize mutational co-occurrence. On the right side is a schematic representation of the mutated pathways. Mutated genes are marked by an arrow, and the prevalence of mutations is reported. The following pathways are shown: NF-κB (A), PI3K/AKT (B), cytokine signaling (C), epigenetics (D), immune surveillance (E), and NOTCH signaling (F).
Pathways that are recurrently mutated in newly diagnosed cHL. In the heat map, rows correspond to genes and columns represent individual patients. Color coding is based on gene alteration status (gray, wild type; red, mutated). The heat map was manually clustered to emphasize mutational co-occurrence. On the right side is a schematic representation of the mutated pathways. Mutated genes are marked by an arrow, and the prevalence of mutations is reported. The following pathways are shown: NF-κB (A), PI3K/AKT (B), cytokine signaling (C), epigenetics (D), immune surveillance (E), and NOTCH signaling (F).
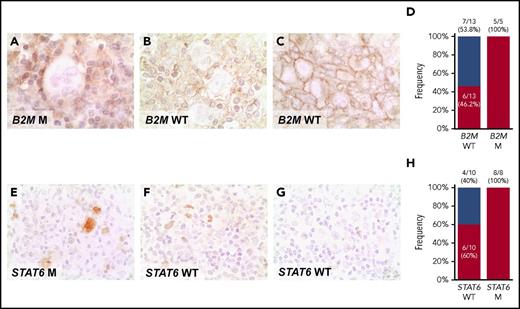
pSTAT6 expression in the nucleus of HRS cells from STAT6 mutated cases provided the proof of principle of the functional consequences of STAT6 mutations (Figure 5E-G). Nuclear pSTAT6 expression by HRS cells was broader than STAT6 mutations in our cHL cohort (Figure 5H), a fact that may be consistent with the plethora of genetic mechanisms affecting the cytokine signaling program in this tumor, some of which were not explored by our assay (eg: SOCS1 mutations and chromosome 9p amplification. B2M mutated cases always lacked MHC-I expression in Hodgkin-Reed Sternberg cells, thus indirectly validating B2M variants discovered in cfDNA (Figure 5A-C). MHC-I was absent in a proportion of B2M wild-type patients (Figure 5D). In those cases, alternative genetic mechanisms not explored by our assay, such as B2M gene locus deletion or mutation/deletion of the HLA genes, may be responsible for the loss of MHC-I expression, as previously reported in DLBCL.28
Expression of phosphorylated STAT6 and MHC-I proteins in cHL. Expression of MHC-I (A-C), and pSTAT6 (E-G) was examined by immunohistochemistry in formalin-fixed sections of cHL. (A) HRS cells with absent expression of MHC-I in a exemplificative B2M mutated cHL case. (B) HRS cells with absent expression of MHC-I in a exemplificative B2M wild-type (WT) cHL case, in which alternative genetic mechanisms not explored by our assay, may be responsible for the loss of MHC-I expression. (C) HRS cells with normal expression of MHC-I in a exemplificative B2M WT cHL case. Staining for MCH-I was absent on the HRS cell surface of 11/18 patients with cHL, as clearly shown by the lack of surface staining at the cell boundaries in the cluster of HRS cells, including 5/5 cases harboring B2M mutations and 6/13 cases lacking B2M mutations (D). (E) HRS cells with a nuclear overexpression of p-STAT6 in a cHL case harboring STAT6 mutation. (F) HRS cells with a nuclear overexpression of p-STAT6 in a exemplificative STAT6 WT cHL case, consistent with the plethora of genetic mechanisms affecting the cytokine signaling program in this tumor, some of which are not explored by our assay and may be responsible for the overexpression of p-STAT6. (G) HRS cells with absent nuclear expression of p-STAT6 in a exemplificative STAT6 WT cHL case. Staining for nuclear pSTAT6 was positive in HRS cells of 14/18 patients with cHL, including 8/8 cases harboring STAT6 mutations and 6/10 cases lacking STAT6 mutations (H). Original magnifications ×630.
Expression of phosphorylated STAT6 and MHC-I proteins in cHL. Expression of MHC-I (A-C), and pSTAT6 (E-G) was examined by immunohistochemistry in formalin-fixed sections of cHL. (A) HRS cells with absent expression of MHC-I in a exemplificative B2M mutated cHL case. (B) HRS cells with absent expression of MHC-I in a exemplificative B2M wild-type (WT) cHL case, in which alternative genetic mechanisms not explored by our assay, may be responsible for the loss of MHC-I expression. (C) HRS cells with normal expression of MHC-I in a exemplificative B2M WT cHL case. Staining for MCH-I was absent on the HRS cell surface of 11/18 patients with cHL, as clearly shown by the lack of surface staining at the cell boundaries in the cluster of HRS cells, including 5/5 cases harboring B2M mutations and 6/13 cases lacking B2M mutations (D). (E) HRS cells with a nuclear overexpression of p-STAT6 in a cHL case harboring STAT6 mutation. (F) HRS cells with a nuclear overexpression of p-STAT6 in a exemplificative STAT6 WT cHL case, consistent with the plethora of genetic mechanisms affecting the cytokine signaling program in this tumor, some of which are not explored by our assay and may be responsible for the overexpression of p-STAT6. (G) HRS cells with absent nuclear expression of p-STAT6 in a exemplificative STAT6 WT cHL case. Staining for nuclear pSTAT6 was positive in HRS cells of 14/18 patients with cHL, including 8/8 cases harboring STAT6 mutations and 6/10 cases lacking STAT6 mutations (H). Original magnifications ×630.
We next compared the ctDNA genetic signatures of cHL, DLBCL, and primary mediastinal large B-cell lymphoma by applying a probabilistic classifier derived from differentially represented mutations. As already shown by gene expression profiling,25 cHL and primary mediastinal large B-cell lymphoma clustered together also genetically, whereas the majority (79.5%) of cHL showed a distinct mutational signature compared with DLBCL (supplemental Figure 7).
Noninvasive monitoring of clonal evolution in refractory cHL
To identify mutations that are enriched in refractory cHL, 32 patients who relapsed after salvage autologous stem cell transplant were genotyped on ctDNA, and their aggregated mutational profile was compared with that of newly diagnosed patients. Paired analysis of HRS cells microdissected from biopsies collected along with plasma at the time of relapse confirmed that ctDNA also mirrors the tumor genotype at this point of the disease (Figure 1E).
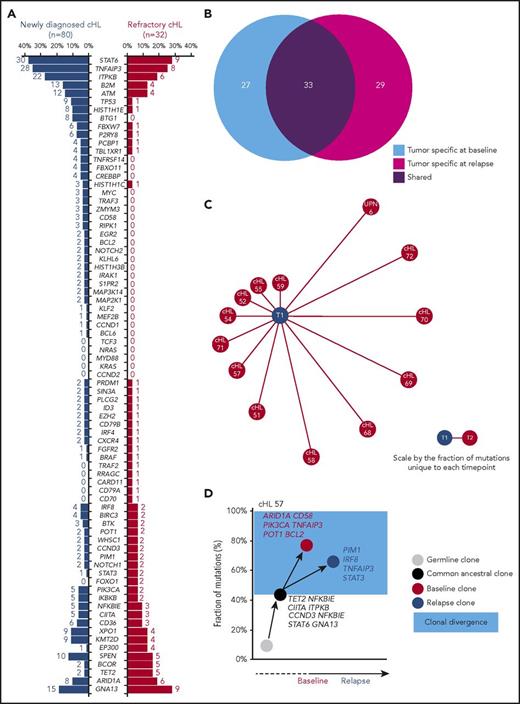
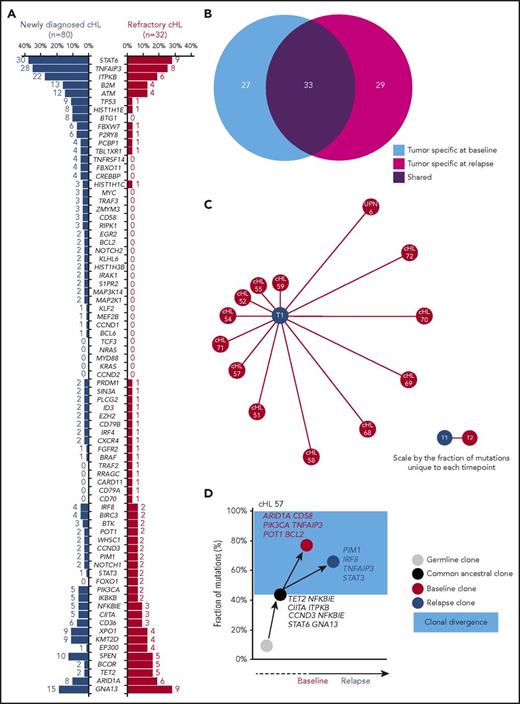
Overall, the mutational profiles of newly diagnosed and refractory cHL were largely overlapping (Figure 6A), suggesting that gene mutations not covered by our target region or molecular mechanisms not captured by CAPP-seq might contribute to the chemorefractory phenotype. The origin of ctDNA mutations from clonal hematopoiesis selected by chemotherapy rather than cHL was ruled out by the observation that they did not occur in paired peripheral blood granulocytes collected at the same time of the plasma sample.29
Genotype and clonal evolution of refractory cHL. (A) Comparison between the mutational profile of newly diagnosed cHL (n = 80; blue bars) vs refractory cHL (n = 32; red bars). Number and prevalence of mutated cases is given for each gene. (B) Venn diagram showing the number of tumor-specific mutations at baseline (light blue circle), the number of tumor-specific mutations at cHL relapse (magenta circle), and the number of tumor mutations shared between the 2 points (purple circle) in longitudinal samples from 13 patients with refractory cHL. (C) Schematic representation of mutational divergence between baseline/relapse pairs of 13 refractory cHL cases. The central node represents baseline (T1), and the distance between baseline and each refractory relapse (edge) is expressed as the fraction of unique mutations to both baseline and relapse points. (D) Phylogenetic tree describing evolutionary distances between sequential tumor pairs in a refractory cHL case. Inferred ancestral clones are also indicated. Evolutionary distance is defined as the fraction of shared mutations between tumor at the baseline and relapse time points (black node), the fraction of mutations specific for the baseline time point (red node), and the fraction of mutations specific for the relapse point (blue node). On the y-axis, 100% represents all mutations identified throughout the longitudinal course of the patient. The time between baseline and relapse is depicted on the x-axis. The leftmost node (gray circle) indicates the germline cell, and the second node from the left (black circle) indicates the last common inferable ancestral clone. The length of the branch between the germline cell and the common ancestral clone reflects the fraction of shared mutations between baseline and relapse samples. The length of the branches between the common ancestral clone and either baseline or relapse samples reflects the fractions of mutations observed only in baseline or relapse samples, respectively. The colored light blue area represents the clonal divergence between baseline and relapse points. Mutated genes shared between baseline and relapse time points (black), mutated genes specific for baseline (red), and mutated genes specific for relapse time point (blue) are annotated.
Genotype and clonal evolution of refractory cHL. (A) Comparison between the mutational profile of newly diagnosed cHL (n = 80; blue bars) vs refractory cHL (n = 32; red bars). Number and prevalence of mutated cases is given for each gene. (B) Venn diagram showing the number of tumor-specific mutations at baseline (light blue circle), the number of tumor-specific mutations at cHL relapse (magenta circle), and the number of tumor mutations shared between the 2 points (purple circle) in longitudinal samples from 13 patients with refractory cHL. (C) Schematic representation of mutational divergence between baseline/relapse pairs of 13 refractory cHL cases. The central node represents baseline (T1), and the distance between baseline and each refractory relapse (edge) is expressed as the fraction of unique mutations to both baseline and relapse points. (D) Phylogenetic tree describing evolutionary distances between sequential tumor pairs in a refractory cHL case. Inferred ancestral clones are also indicated. Evolutionary distance is defined as the fraction of shared mutations between tumor at the baseline and relapse time points (black node), the fraction of mutations specific for the baseline time point (red node), and the fraction of mutations specific for the relapse point (blue node). On the y-axis, 100% represents all mutations identified throughout the longitudinal course of the patient. The time between baseline and relapse is depicted on the x-axis. The leftmost node (gray circle) indicates the germline cell, and the second node from the left (black circle) indicates the last common inferable ancestral clone. The length of the branch between the germline cell and the common ancestral clone reflects the fraction of shared mutations between baseline and relapse samples. The length of the branches between the common ancestral clone and either baseline or relapse samples reflects the fractions of mutations observed only in baseline or relapse samples, respectively. The colored light blue area represents the clonal divergence between baseline and relapse points. Mutated genes shared between baseline and relapse time points (black), mutated genes specific for baseline (red), and mutated genes specific for relapse time point (blue) are annotated.
To define cHL clonal evolution patterns, we genotyped longitudinal ctDNA samples (n = 41 from 13 patients) collected before frontline treatment (ie, ABVD), at the time of relapse, and during salvage therapies comprising transplantation, brentuximab vedotin, and nivolumab. The amount of input cfDNA for library preparation and sequencing coverage were similar across longitudinal samples, indicating that differences in the representation of mutations was not the result of a difference in the sensitivity of the CAPP-seq (supplemental Figure 2C-D). Clonal shifts between pretreatment and relapse samples were documented in all cases (Figure 6B-C), thus demonstrating that evolution after therapy is the rule. In patients relapsing under/after chemotherapy or brentuximab vedotin, diagnosis/relapse tumor pairs branched through the acquisition of phase-specific mutations from an ancestral clone that persisted throughout disease course (Figure 6D; supplemental Figure 8). Mutations of STAT6, GNA13, ITPKB, and TNFAIP3 were preferentially inferred in the ancestral clones, indicating that they are an early event in cHL (Figure 6D; supplemental Figure 8). Overall, these data point to ctDNA genotyping as a tool for the noninvasive monitoring of cHL clonal evolution and suggest that in a refractory setting, chemotherapy and chemotoxins only partially reshape the subclonal composition, leaving intact the ancestral clones.
Among patients maintaining a partial response under nivolumab, ancestral clones were cyclically suppressed and replaced by novel clones harboring new mutations (supplemental Figure 9). Such a pattern might be interpreted as a drug-promoted aggression against cancer neoantigens stemming from mutations and the attempt made by the tumor to evade treatment by generating new mutations.
Dynamics of circulating tumor DNA during therapy complements dynamics of PET/CT in cHL response assessment
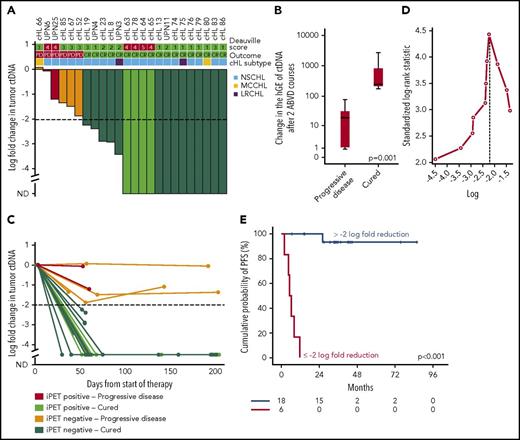
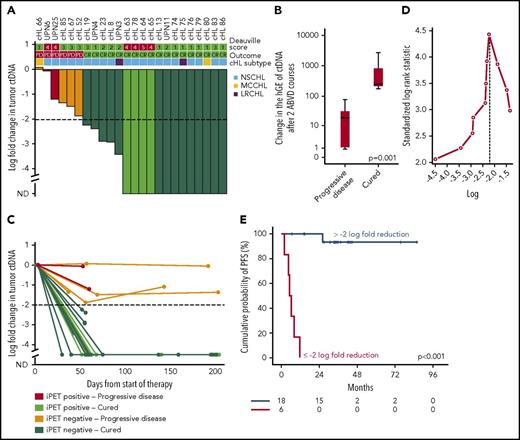
We followed serial plasma samples during therapy in a cohort of 24 patients with advanced cHL treated with ABVD. Patients achieving complete response and cure had a larger drop in ctDNA load after 2 ABVD courses compared with relapsing patients (Figure 7A-B), and the magnitude of the drop was maintained until the end of the therapy (Figure 7C). A drop of 100-fold or 2-log drop in ctDNA after 2 chemotherapy courses, a threshold proposed and validated in DLBCL30 and confirmed as best cutoff to predict progression in our cohort (Figure 7D), was associated with complete response and cure (Figure 7A,E). Conversely, a drop of less than 2-log in ctDNA after 2 ABVD courses was associated with progression and inferior survival (Figure 7A,E; supplemental Figure 10). Quantification of ctDNA complemented interim PET/CT in determining residual disease. Indeed, cured patients who were inconsistently judged as interim PET/CT-positive had a more than 2-log drop in ctDNA, whereas relapsing patients who were inconsistently judged as interim PET/CT negative had a less than 2-log drop in ctDNA (Figure 7A). The lack of correlation between log fold change of maximum standardized uptake value and log fold change of ctDNA between baseline and interim evaluations (supplemental Figure 11) is consistent with the notion that maximum standardized uptake value largely reflects the metabolic activity of the inflammatory component of the mass in cHL, whereas ctDNA reflects the tumor load levels.
Change in tumor ctDNA is a prognostic biomarker in cHL treated with chemotherapy. (A) Waterfall plot of the log-fold change in ctDNA load after 2 courses of ABVD in 24 advanced stage cHL cases. On top of the graph, the interim PET/CT response scored according to the Deauville criteria and the final outcome of the patient are indicated. Histological subtype of cHL are shown above the plot. Each column is color coded according to the interim PET/CT results and the final patient outcome. Levels of ctDNA are normalized to baseline levels. The dash line tracks the −2-log threshold (CR, complete remission and cure; iPET, interim PET/CT; ND, not detectable; PD, progressive disease). (B) Box plot showing the fold change of ctDNA after 2 ABVD courses among patients who progressed vs patients who were cured. The band inside the box is the median, the bottom and top of the box are the first and third quartiles, and the ends of the whiskers represent the range. P value by Mann-Whitney U test. (C) Spider plot showing the log-fold change in ctDNA load after 2 courses of ABVD and at the end of therapy. Dots represent individual ctDNA measurements at the specified points. Each line is color coded according to the interim PET/CT results and the final patient outcome. (D) Maximally selected log-rank statistics identifies ∼2-log-fold reduction in ctDNA levels after 2 ABVD courses as the best cutoff for progression-free survival (PFS) anticipation. (E) Kaplan-Meier curve of PFS stratified according to whether a 2-log reduction in ctDNA was achieved or not after 2 ABVD courses in 24 advanced stage cHL cases. Among patients achieving more than 2-log reduction after 2 ABVD courses, the sole event registered was a death in remission. Conversely, among patients achieving less than 2-log reduction after 2 ABVD courses, all the events were progressions.
Change in tumor ctDNA is a prognostic biomarker in cHL treated with chemotherapy. (A) Waterfall plot of the log-fold change in ctDNA load after 2 courses of ABVD in 24 advanced stage cHL cases. On top of the graph, the interim PET/CT response scored according to the Deauville criteria and the final outcome of the patient are indicated. Histological subtype of cHL are shown above the plot. Each column is color coded according to the interim PET/CT results and the final patient outcome. Levels of ctDNA are normalized to baseline levels. The dash line tracks the −2-log threshold (CR, complete remission and cure; iPET, interim PET/CT; ND, not detectable; PD, progressive disease). (B) Box plot showing the fold change of ctDNA after 2 ABVD courses among patients who progressed vs patients who were cured. The band inside the box is the median, the bottom and top of the box are the first and third quartiles, and the ends of the whiskers represent the range. P value by Mann-Whitney U test. (C) Spider plot showing the log-fold change in ctDNA load after 2 courses of ABVD and at the end of therapy. Dots represent individual ctDNA measurements at the specified points. Each line is color coded according to the interim PET/CT results and the final patient outcome. (D) Maximally selected log-rank statistics identifies ∼2-log-fold reduction in ctDNA levels after 2 ABVD courses as the best cutoff for progression-free survival (PFS) anticipation. (E) Kaplan-Meier curve of PFS stratified according to whether a 2-log reduction in ctDNA was achieved or not after 2 ABVD courses in 24 advanced stage cHL cases. Among patients achieving more than 2-log reduction after 2 ABVD courses, the sole event registered was a death in remission. Conversely, among patients achieving less than 2-log reduction after 2 ABVD courses, all the events were progressions.
Immune checkpoint inhibitors can cause false-positive PET/CT results in cHL as a result of tumor flares or pseudoprogression.17 We followed longitudinal plasma samples collected during therapy with nivolumab from 5 patients with refractory cHL. Among 4 patients stabilized in partial remission, ctDNA load did not change under nivolumab treatment, thus reflecting the persistence of the tumor (supplemental Figure 9). One patient achieved a PET/CT complete remission with clearance of ctDNA. He became ctDNA-positive during a PET/CT, and biopsy confirmed tumor flare (supplemental Figure 1E).
Discussion
The study establishes ctDNA as source of tumor DNA for cHL mutational profiling. By overcoming the major technical hurdles that have so far limited cHL genotyping, our technical approach based on ctDNA has allowed large-scale assessment of mutations in different clinical phases of the disease, ranging from newly diagnosed to refractory disease, and longitudinally during disease treatment.
Despite targeted resequencing being underpowered compared with exome sequencing in discovering new candidates, this study refines the current knowledge of cHL genetics. First, STAT6 is identified as the most frequently mutated gene in cHL, which is in keeping with the known importance of cytokine signaling in the biology of this tumor,2,13,25 but was not reported in previous exome sequencing studies of cHL cases.19 Second, mutation recurrence of previously discovered genes is defined more precisely. As an example, our approach has narrowed the estimate of the recurrence of B2M mutations in cHL, which is lower than previously reported,19 and consistent with that observed in another cHL study.31 Third, the notion that different histologic subtypes of cHL are biologically distinct, as previously shown at the transcriptome level,25 is extended and reinforced at the genetic level.19,20 Fourth, a few major pathways emerged as recurrently mutated, including NF-κB, PI3K-AKT, cytokine and NOTCH signaling, and immune evasion. Of note, these pathways have been previously identified by gene expression profiling and functional genomic studies of cHL,13,21,22,24,25,32 indicating that mutations act as red flags highlighting cellular programs that are relevant for the biology of the disease and potential therapeutic targets.
By longitudinally profiling patients with cHL treated with ABVD or brentuximab vedotin, we provide the evidence for a reservoir ancestral population that, as in other lymphoma models,33 is resistant to chemotherapy and propagates successive disease relapse. New mutations appearing in ctDNA mark resistant clones that are selected during the clonal evolution process taking place under the selective pressure of treatment, although they may not be directly causative of relapse.
Immunotherapy is conversely capable of eradicating ancestral clones, and its selective pressure elicits a quick clonal evolution that was unexpected in cHL, resulting in the complete reshape of the disease mutation profile in few months. The evolving landscape of mutations, which are the major source of neoantigens in cancer,34 is a mechanism of tumor escape from immunotherapy35,36 and might be at the basis of either the long-lasting partial responses or periodical tumor flares that patients typically experience under treatment with immune checkpoint blockers.37,38
Although interim PET/CT response assessment is a novel approach to refine management strategies before completing treatment in cHL,39-41 meta-analyses demonstrated a certain degree of inaccuracy of this application.42,43 To fill this gap, an area of growing interest is pairing interim PET/CT with biomarkers to enhance their cumulative predictive value.44,45 Our results provide the proof of principle that ctDNA can measure residual disease during treatment in cHL. Moreover, our data generated the hypotheses that ctDNA quantification after 2 chemotherapy courses may have prognostic implications, and that ctDNA may complement interim PET/CT in informing on patients’ outcome. Incorporation of both PET/CT and ctDNA monitoring into clinical trials should allow us to precisely define their cumulative sensitivity and specificity in anticipating the clinical course of patients with cHL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by Grant No. KFS-3746-08-2015, Swiss Cancer League, Bern, Switzerland; Grant No. 320030_169670/1, Swiss National Science Foundation, Bern, Switzerland; iCARE No. 17860, Associazione Italiana per la Ricerca sul Cancro and Unione Europea, Milan, Italy; Fondazione Fidinam, Lugano, Switzerland; Nelia & Amadeo Barletta Foundation, Lausanne, Switzerland; Fondazione Ticinese per la Ricerca sul Cancro, Bellinzona, Switzerland; Grant No. RF2011-02346986, Ministry of Health, Rome, Italy; Grant No. 16722 and Grant No. 20575, Italian Association for Cancer Research, Milan, Italy; Fondazione Regionale per la Ricerca Biomedica, Milan, Italy; Fondazione Umberto Veronesi, Milan, Italy; Grant No. 10007, Special Program Molecular Clinical Oncology 5 x 1000, Associazione Italiana per la Ricerca sul Cancro, Milan, Italy; Grant No. RF-2011-02349712, Ministero della Salute, Rome, Italy; and Grant No. 2015ZMRFEA_004, MIUR-PRIN, Rome, Italy.
Authorship
Contribution: D.R. designed the study, interpreted data, and wrote the manuscript; V.S. and A.B. performed molecular studies and bioinformatics analysis and contributed to data interpretation and manuscript preparation; A. Cuccaro, A. Cocomazzi, A. Condoluci, S.L.L., E.C., A.A.M., A. Stathis, L.N., C.D., F.D., R.B., and B.G. provided study material and clinical data and contributed to manuscript revision; A.A. and L.T.-D.-B. contributed to bioinformatics analysis and manuscript preparation; M.D.T., G.F., M. Manzoni, F.G., and A. Cuccaro contributed to sample biobanking, molecular studies, and manuscript revision; M. Martini and L.M.L. performed pathological revision, immunohistochemical studies, microdissections, and manuscript revision; L.C., S.A., V.R., and A.G. contributed to imaging revision and manuscript revision; A.N., F.B., M.G., G.S., A. Santoro, F.C., E.Z., and G.G., contributed to data interpretation and manuscript revision; and S.H. and C.C.-S. provided key scientific insights and reagents and contributed to data interpretation and manuscript revision.
Conflict-of-interest disclosure: D.R. received unrestricted research grants from Gilead and AbbVie and honoraria from Gilead, AbbVie, and Janssen. A. Santoro received honoraria from Merck Sharp & Dome, Bristol, and Roche. The remaining authors declare no competing financial interests.
Correspondence: Davide Rossi, Hematology, Oncology Institute of Southern Switzerland and Institute of Oncology Research, 6500 Bellinzona, Switzerland; e-mail: davide.rossi@eoc.ch.
References
Author notes
V.S. and A.B. contributed equally to this study.
S.H., C.C.-S., and D.R. contributed equally to this study.