Abstract
Introduction: DLBCL has two COO subtypes: Activated B Cell (ABC) and Germinal Center B cell (GCB). Patients with the ABC subtype have a poor prognosis compared to those with GCB, and COO can be predictive for response to some new therapeutic agents. Traditionally, COO subtype has been determined by microarray (ABC, GCB, unclassified), IHC-based algorithms (GCB or non-GCB), or expression-based Nanostring Lymph2Cx (ABC, GCB, unclassified). Some reports have failed to show a prognostic difference between GCB and non-GCB when employing IHC-based algorithms. This has led some to adopt the Lymph2Cx assay as the preferred method to assess COO, but in some cases the tumor content or RNA quality is inadequate to perform this assay. COO subtypes have differing gene mutations, with GCB typically characterized by EZH2 alterations and IGH:BCL2 translocations, while ABC is dominated by NF-KB and BCR signaling alterations such as MYD88 and CD79B short variants. Here we utilized mutational differences in COO subtypes to develop a COO DNA classification (COODC) model to predict COO from DNA-based features on a clinically utilized platform.
Methods: Comprehensive genomic profiling (CGP) of DLBCL samples was performed using the DNA component of the FoundationOne® Heme platform; sequencing 465 genes for the GOYA trial (R-CHOP vs G-CHOP; N=499; NCT01287741), MAIN trial (R-CHOP +/- bevacizumab; N=44; NCT00486759), and cases from routine clinical care (FM-clinical; N=597). Gold standard COO classifications were determined in GOYA using the Lymph2Cx assay, and in MAIN using a modified Wright algorithm applied to a custom Nanostring expression panel. COODC was developed using a penalized lasso regression with 25-fold internal cross validation and cutoffs that optimize specificity and sensitivity. Concordance was calculated as the percentage of COODC calls matching the gold standard, excluding samples called unclassified in the gold standard or COODC.
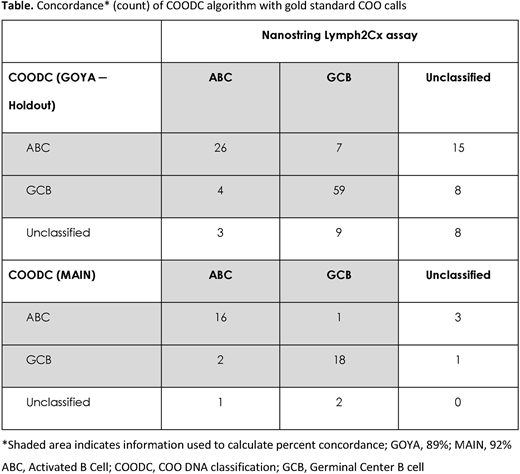
Results: We developed a novel DNA-based method to determine COO, which is 89% concordant with Lymph2Cx (Table) COODC was trained on 296 GOYA samples with Lymph2Cx COO calls and validated on 139 held-out samples from the same cohort (GOYA). Additional validation was performed on an independent first-line cohort (MAIN) and confirmed a high concordance (92%). COODC also provides prognostic value, confirming the worse prognosis of ABC (progression-free survival [PFS] hazard ratio [HR] 1.6; 95% confidence interval [CI]: 1.1-2.4; p=0.011) and unclassified (PFS HR 1.9; 95% CI: 1.1-3.2; p=0.021) compared with GCB. This approach also allows investigation into the genomics and underlying features of COO subtypes. Alterations in BCL2, EZH2, and TNFRSF14 were important determinants of the GCB phenotype, consistent with the enrichment of GCB samples among the EZB cluster in Schmitz et al. (N Engl J Med 2018), or C3 in Chapuy et al. (Nat Med 2018). Alterations in MYD88 and CD79B were highly predictive of ABC subtype, as expected. Alterations in NOTCH1, NOTCH2, and BCL6 were all slightly predictive of ABC subtype, despite the mixed phenotype of the BN2, N1 (Schmitz et al. N Engl J Med 2018), and C1 (Chapuy et al. Nat Med 2018) groups. Novel features include decreased average copy number of chromosome 6p, which encodes the HLA locus, in ABC (p=0.0064), and increased frequency of T>A and T>G alterations in GCB (p<0.0001 for both). We identified 2 major mutational signatures associated with COO: COSMIC signature 23, corresponding to a canonical activation-induced cytidine deaminase signature, enriched in ABC (p=0.002), and COSMIC signature 3, which trends to increased frequency in GCB (p=0.13).
Conclusions: We have developed a new and clinically relevant method for determining DLBCL COO in specimens with tumor purity as low as 20%, without the need for RNA or matched normal tissue. This method is 89% (GOYA) to 92% (MAIN) concordant with the Lymph2Cx assay and maintains prognostic value. Integrating COO with CGP enables insights into disease biology, including the identification of novel features associated with COO. We identified a difference in mutational signatures between ABC and GCB, suggesting that different origin cells may contribute to different mutational processes.
Trabucco:Foundation Medicine Inc: Employment. Sokol:Foundation Medicine: Employment. Moore:Foundation Medicine, Inc.: Employment. Maund:Genentech: Employment. Frampton:Foundation Medicine Inc: Employment, Other: Employee and stockholder. Miller:Foundation Medicine: Employment, Other: Ownership interests none PLC*; Revolution Medicines: Membership on an entity's Board of Directors or advisory committees. Venstrom:Genentech Inc: Employment. Albacker:Foundation Medicine Inc: Employment; Foundation Medicine Inc: Employment. Sehn:Morphosys: Consultancy, Honoraria; Seattle Genetics: Consultancy, Honoraria; Karyopharm: Consultancy, Honoraria; Celgene: Consultancy, Honoraria; Lundbeck: Consultancy, Honoraria; TG Therapeutics: Consultancy, Honoraria; Merck: Consultancy, Honoraria; Roche/Genentech: Consultancy, Honoraria; Abbvie: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Janssen: Consultancy, Honoraria. Oestergaard:Roche: Employment, Other: Ownership interests PLC. Bolen:Roche: Other: Ownership interests PLC*.
Author notes
Asterisk with author names denotes non-ASH members.