Abstract
Background: Five to ten percent of patients with sickle cell disease (SCD) have a high risk of suffering an ischemic stroke before they are 10 years of age. The clinical consequences of stroke are severe yet our current mechanistic understanding of the causes of stroke in SCD is limited. There is strong evidence that there is genomic contribution to the risk of stroke, other than just the SCD gene mutation. By whole exome sequencing, we have shown that a specific coding variant in the GOLGB1 gene, Y1212C, is significantly associated with decreased risk of stroke. GOLGB1 encodes a conserved Golgi membrane protein that is essential for maintaining Golgi structure and associated protein transport. To elucidate how the GOLGB1 Y1212C variant is protective against stroke risk in SCD, we performed morphological analysis of Golgi in cultured primary macrophages and whole proteome analyses of peripheral immune cells isolated from SCD patients.
Methods: For primary macrophage preparations, we collected whole blood from non-SCD controls (n=11) and pediatric SCD patients (n=12). Monocytic CD14 positive cells were isolated and cultured for nine days with granulocyte monocyte stimulating colony factor, fixed and sequentially stained with antibodies against GOLGB1, TGN-46 and GM130. DAPI was used as a nuclear stain. High resolution images were acquired on Zeiss LSM 780 laser scanning confocal microscope with a 63X oil objective. Golgi morphological analysis was performed using a MATLAB script for cell segmentation, estimation of individual cell area, quantification of Golgi fragments per cell, and Golgi volume and organelle spread index. For the proteomic analysis, we isolated peripheral blood mononuclear cells from 7 patients with SCD. These cells were immediately flash-frozen and stored at -80oC until proteomic processing. We used a label-free protocol using dual-pH reverse phase fractionation and peptide sequencing of digested samples on a Thermo Scientific Orbitrap mass spectrometer. Protein abundances in each sample were estimated by using the intensity-based absolute quantification algorithm and total protein normalization was done with a fraction of total score (iFOT). Data were evaluated using GraphPad Prism and statistical analyses for differences between SCD and non-SCD samples or between samples with and without the GOLGB1 variant were performed with unpaired Student's t-test or Kruskal-Wallis test; analyses for multiple group comparisons were performed with the one-way ANOVA method. Statistical significance was set at P < 0.05.
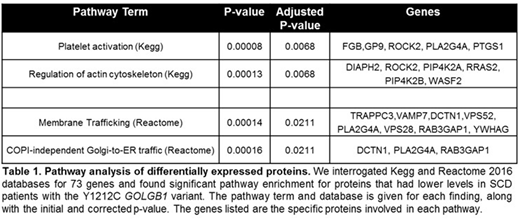
Results: In cultured primary macrophages, SCD individuals with the GOLGB1 Y1212C variant had altered Golgi morphologies compared to individuals with wild-type GOLGB1. Overall, macrophages derived from patients with SCD (n=12) were significantly larger (average cell size 11,728 vs. 7,598; p<0.001) yet had smaller Golgi (average Golgi volume 69.6µm vs. 75.4µm, p<0.01) and with more detected Golgi fragments (12.3 vs. 10.2 per cell, p<0.01) compared to non-SCD controls (n=11). For SCD patients with the GOLGB1 variant (n=5 heterozygote and 1 homozygote), their Golgi apparatus were more compact (average spread index 0.087 vs. 0.079; p<0.05) and had less Golgi fragments per cell (average 10.9 vs. 13.9; p<0.05) compared to SCD patients wild-type for GOLGB1 (n=6). From the proteomics analysis of peripheral blood immune cells, we identified 73 proteins that were differentially expressed (p<0.05) between SCD patients with (n=3) and without (n=4) the GOLGB1 variant. Of these proteins, 64 (90%) had lower levels in patients with the GOLGB1 Y1212C variant. Pathway analysis of the identified proteins showed that there strong clustering of proteins involved in platelet activation, regulation of actin cytoskeleton, and COPI-independent Golgi-to-ER retrograde trafficking (Table 1).
Conclusions: Our study has shown that a coding variant in GOLGB1, identified as protective against risk of stroke in patients with SCD, has significant effects on Golgi function in SCD samples. We observed that having the GOLGB1 Y1212C variant resulted in more compact and less fragmented Golgi apparatus. Proteomic analysis showed that SCD patients with the GOLGB1 variant also had significantly lower levels of proteins involved in platelet activation and Golgi trafficking. Our findings suggest a novel role for the Golgi apparatus in controlling protein flux that modulates risk of stroke in SCD.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.