Abstract
Introduction: Heparin-induced thrombocytopenia (HIT) is a complication of heparin exposure wherein antibodies (primarily IgG) specific for heparin/platelet-factor-4 (H/PF4) superstructures lead to platelet activation, consumptive thrombocytopenia, and acquired thrombophilia. The immune response is presumably elicited by exposure of a cryptic antigen when heparin and PF4 are bound. However, neither heparin nor PF4 is a foreign antigen in humans, raising the question whether loss of self-tolerance via aberrant antigen presentation by human leukocyte antigen (HLA) may contribute to the development of H/PF4 antibodies. We present evidence that development of high H/PF4 antibody levels is associated with specific HLA alleles, identification of which may elucidate mechanisms for antibody origin and principles for HIT risk stratification.
Methods: We conducted a case-control study of 412 adult inpatients tested for enzyme-linked immunosorbent assay H/PF4 antibodies (Diagnostic Stago Asserachrom HPIA-ELISA) for HIT workup after having received intravenous unfractionated heparin. Positive cases (n=60) were individuals with assay optical density (OD) >1.00 while negative controls (n=352) had OD ≤1.00. DNA extracted from peripheral blood of cases and controls were sequenced at HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 loci (Illumina). High-resolution HLA typing results were analyzed for association with case/control status by Fisher's exact test.
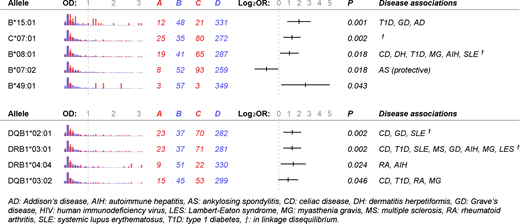
Results: Median (IQR) OD was 0.16 (0.08-0.34) for controls and 1.68 (1.23-2.31) for cases. The highest case OD was 3.00. Several HLA alleles were associated with case/control status (Figure 1). For class I loci, B*15:01 (odds ratio, OR: 3.94; 95%CI: 1.82-8.52), C*07:01 (2.43; 1.37-4.30), B*08:01 (2.05; 1.12-3.75), and B*49:01 (6.12; 1.21-31.08) were risk factors for developing H/PF4 antibodies, while B*07:02 (0.43; 0.20-0.94) was protective. For class II loci, DQB1*02:01 (2.50; 1.40-4.48), DRB1*03:01 (2.46; 1.37-4.40), DRB1*04:04 (2.65; 1.15-6.07), and DQB1*03:02 (1.88; 0.98-3.61) were risk factors. After adjusting for age, gender, race, and multiple comparison, B*15:01, C*07:01, DQB1*02:01, and DRB1*03:01 remained significant risk factors (p <0.05). Linkage disequilibrium was present between B*08:01, C*07:01, DQB1*02:01, and DRB1*03:01.
Discussion: We identified several HLA alleles associated with H/PF4 antibody formation, comprised almost entirely of well-known risk alleles for autoimmune disease. DQB1*02:01 (DQ2) and DQB1*03:02 (DQ8) are present in nearly all cases of celiac disease. DRB1*03:01 (DR3), linked with DQ2, is the single HLA allele most frequently associated with autoimmune diseases in the literature. B*08:01 (B8) is part of the "super B8" haplotype overlapping with DR3. B*15:01 (B15) has reported associations with type 1 diabetes, Grave's disease and Addison's disease. B*08:01, C*07:01, DQB1*02:01, and DRB1*03:01 appeared to constitute a distinct risk haplotype, separate from B*15:01 or DQB1*03:02. We also identified a protective allele, B*07:02, which has protective associations with ankylosing spondylitis.
Our findings suggest individuals who develop high H/PF4 antibody levels following heparin exposure share HLA haplotypes with individuals who develop auto-antibodies in other contexts, perhaps via shared pathophysiology. A history of autoimmune disease may increase one's pre-test probability for HIT compared to the general population, and merits consideration when HIT is on the differential diagnosis. With additional outcomes research, such history might form a useful adjunct to the "4T" screen. Functional implications of HLA associations with HIT warrant further investigation.
Figure 1. Human leukocyte antigen alleles associated with heparin/platelet-factor-4 antibody formation. Shown are histograms of optical density (OD) value, contingency-table counts for cases or controls having or missing an allele, odds ratios (OR) in base-2 logarithmic scale with 95% confidence interval, one-tailed Fisher's exact p-value, and known disease associations for each allele. Red: allele present, blue: allele absent, A: cases having allele, B: cases missing allele, C: controls having allele, D: controls missing allele.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.