In this issue of Blood, Beauchamp et al1 reveal that the kinase CDK9, typically associated with transcriptional elongation,2 directly modulates translation through coopting components of the major translational regulatory complex mammalian target of rapamycin (mTOR).3,4 These studies represent a major shift from the current paradigm; specifically, this work demonstrates that other kinases can coopt mTOR cofactors in order to modulate not only translation but also transcription as well as positioning CDK9 as an RNA processing cofactor.
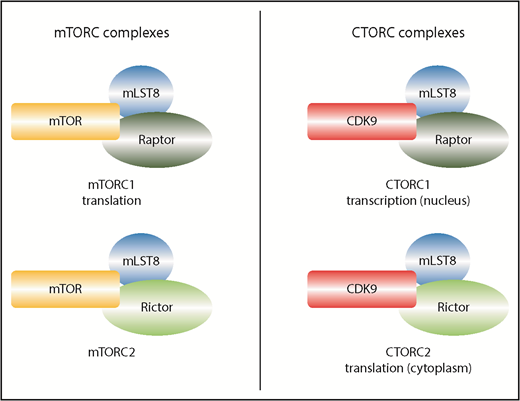
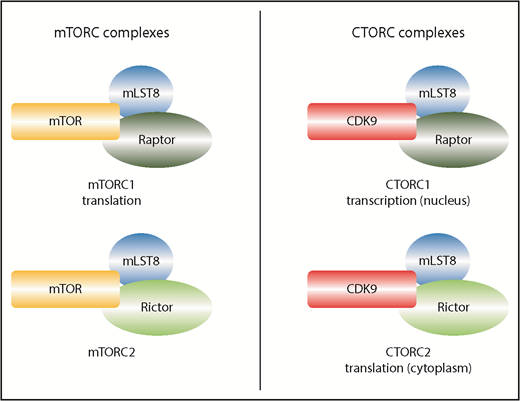
Comparison of mTOR and CDK9 complexes. mTOR is largely cytoplasmic, whereas CDK9 is found in both compartments. To reduce complexity, only the most salient cofactors are shown. mLST8 and RAPTOR interact with mTOR to form mTORC1 and separately with CDK9 to form CTORC1. Although the composition is very similar, the functions of these 2 complexes are distinct, with mTORC1 acting in translation in the cytoplasm and CTORC1 acting in transcription in the nucleus. mLST8 and RICTOR interact with mTOR to form mTORC2 and with CDK9 to form CTORC2. Again, these complexes have different functions, with CTORC2 modifying the overall ribosome profile, whereas mTORC2 is not considered to act in translation.
Comparison of mTOR and CDK9 complexes. mTOR is largely cytoplasmic, whereas CDK9 is found in both compartments. To reduce complexity, only the most salient cofactors are shown. mLST8 and RAPTOR interact with mTOR to form mTORC1 and separately with CDK9 to form CTORC1. Although the composition is very similar, the functions of these 2 complexes are distinct, with mTORC1 acting in translation in the cytoplasm and CTORC1 acting in transcription in the nucleus. mLST8 and RICTOR interact with mTOR to form mTORC2 and with CDK9 to form CTORC2. Again, these complexes have different functions, with CTORC2 modifying the overall ribosome profile, whereas mTORC2 is not considered to act in translation.
In particular, Beauchamp et al, employing proteomics strategies, discovered that CDK9 interacted with mLST8, a component of mTORC1 and mTORC2 complexes and that this was independent of the kinase mTOR. Conventionally, mTORC1 complexes regulate ribosomal biogenesis and translation (among other things), whereas mTORC2 complexes are usually associated with cytoskeletal rearrangement, glucose metabolism, as well as others3 (see figure). Although mLST8 is an important core factor, its knockout does not phenocopy the mTOR knockout,5 hinting that it could have functions beyond mTOR. Here, Beauchamp et al also demonstrate that CDK9 binds not only mLST8 but also other proteins that can be found in the mTOR complexes (eg, RAPTOR and RICTOR). The authors dub these CDK9-containing assemblies CTORC complexes (see figure).
The authors dissected the relevance of these complexes to the nuclear and cytoplasmic activities of CDK9. In the nucleus, they show that CDK9 forms a complex with mLST8, and RAPTOR (referred to at CTORC1) at promoters, strongly indicating that these traditional mTOR components could also act in transcription. The cytoplasmic studies also revealed exciting insights. Although CDK9 traditionally acts in transcription, these studies revealed that it also interacted with a wide range of proteins involved in translation, RNA processing, RNA localization, splicing, etc. Indeed, the cytoplasmic CTORC2 complex (comprising RICTOR, mLST8) is associated with and/or impacted on the phosphorylation of proteins that are functionally associated with the ribosomes. To investigate if CTORC2 modified translation, the authors investigated the formation of polyribosomes (“polysomes”) as a function of CDK9 activity in acute myeloid leukemia (AML) cell lines. Indeed, the authors demonstrate that inhibition of CDK9 with Atuveciclib significantly reduced the formation of polysomes, which would suggest reduced translational efficiency, at least for some transcripts. A combination of mTOR inhibitors, such as Rapamycin or Vistusertib, with Atuveciclib led to the most pronounced impact on polysomes. Although the authors did not observe changes in translation efficiency for the few RNAs interrogated, this striking change to the polysome profile suggests that a good many transcripts will have their translation modulated by CTORC2. It will be an important avenue of research to determine the RNAs that are specifically targeted, likely by carrying out RNA-Seq of polysomal fractions or similar strategies.
Leveraging their new-found biochemical insights, the authors explored the effects of inhibiting CDK9 with Atuveciclib in in vivo, ex vivo, and in vitro models of AML. In these cases, they compared its activity alone with that of the cornerstone of AML therapy, cytarabine. The combination of cytarabine and Atuveciclib yielded the most robust responses. Atuveciclib is currently being tested in a phase 1 clinical trial in advanced leukemia patients, suggesting the insights garnered here could be rapidly translated into the clinic (ClinicalTrials.gov, #NCT02345382). These studies also provide important insights as to why targeting mTORC1/2 in the clinic has yielded limited clinical efficacy.4,6
Many exciting questions arise, and new areas of exploration will emerge based on these elegant studies. For instance, CDK9 inhibition alters the association of specific ribosomal proteins with polysomes. Could this mean that CDK9 plays some role in the formation of specialized ribosomes (ie, ribosomes that are optimized for the translation of a certain subset of transcripts)? Oftentimes, factors that play roles in multiple steps of RNA metabolism can coordinate the protein expression of subsets of transcripts that act in the same biochemical pathways, in this way modulating RNA regulons.7 Could CDK9 affect the production of groups of RNAs at the transcription, translation, and perhaps other levels, such as splicing or nuclear export? In this way, could CDK9 be a central node in an RNA regulon that supports malignancy? From the perspective of mTOR components, it will be interesting to understand how many other kinases can coopt these factors and what variety of processes these can function in.
These studies further highlight the importance of RNA processing in AML and other cancers. Although the conventional view has been that dysregulated transcription and signaling are the drivers of cancer, it is clear that dysregulated RNA processing in its many forms (eg, RNA trafficking, translation, stability, splicing, etc) contributes to the oncogenic phenotype and is targetable in malignancies.8,9 AML has already been characterized to have dysregulated RNA processing, including elevated export of RNAs that support malignancy, increased translation, and dysregulated splicing, and these processes can be targeted in patients corresponding to clinical benefit.8-10 The report by Beauchamp et al provides novel insights into the mechanisms that can dysregulate translation and transcription in AML and provides means to exploit these to identify next-generation strategies to target these processes in the clinic.
Conflict-of-interest disclosure: The author declares no competing financial interests.