The outcomes of lymphoma patients treated with PD1 inhibitors (PD1i) vary significantly across different histologies with objective responses rates that are around 70% in classical Hodgkin lymphoma, but significantly lower, around 10%, in indolent non-Hodgkin lymphomas (iNHL). This may be due in part to the inability of PD1i to restore the function of exhausted T-cells (Tex) in the tumor microenvironment (TME). PD1 is expressed on various T-cell subsets, including effector cells and Tex. In this study we sought to identify markers of exhaustion beyond PD1 that define T-cell subsets with high reactivation potential in marginal zone lymphoma (MZL).
To define different stages of exhaustion in MZL, we developed and validated a mass cytometry (CyTOF) panel which integrates T-cell markers of lineage, differentiation, activation, suppression, cytokine production and transcription factors associated with exhaustion. We analyzed 35 MZL spleens obtained at diagnosis and 6 reactive spleens. We identified a population of CD8 cells in MZL that express TIM3 without PD1, compose 1.5% (range 0.14-4.88%) of total CD8 cells and expand with activation through the TCR (6.2% of CD8, range 0.6-14.2%). Prolonged cytokine exposure has been shown to induce exhaustion in healthy T-cells, and so to determine whether PD1-TIM3+ cells are Tex, we treated healthy donor T-cells with activating beads and IL12 and analyzed them by flow cytometry. We showed that PD1-TIM3+ cells expand with repetitive activation and IL12 treatment suggesting that they are Tex. Since Tex have been shown to have a distinct transcription profile compared to functional effector T-cells, we analyzed the expression of several transcription factors (TFs) that are upregulated in Tex by CyTOF and compared them to other subsets of T-cells. PD1-TIM3+ cells expressed higher levels of Eomes, Tbet, TOX and Helios compared to activated effector cells, confirming that they are Tex, but lower levels levels of the same TFs compared to PD1+TIM3+ cells, consistent with a less severely exhausted phenotype.
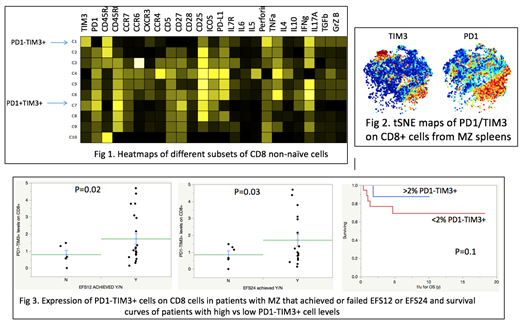
To characterize the function of PD1-TIM3+ cells, we manually gated on subsets of CD8+ non-naïve cells that were stained with the CyTOF panel and compared the different subsets. We showed that PD1-TIM3+ cells arise from effector cells, maintain their polyfunctionality by producing cytokines and chemokines and have a unique cytokine production profile, including perforin, granzyme B and proinflammatory cytokines, such as IL17A. Importantly, PD1-TIM3+ cells produced higher levels of most cytokines compared to more severely Tex expressing both PD1 and TIM3 (Fig 1). We then performed tSNE analysis where similar objects are modeled by nearby points. We determined that PD1-TIM3+ cells cluster together, suggesting that they are phenotypically homogeneous cells in terms of function and expression of activation markers and are significantly different from PD1+TIM3+ cells (Fig 2). We finally blocked PD1 or TIM3 and analyzed the effect of each antibody blockade on function. We showed that TIM3 blockade was able to restore the production of most cytokines to higher degrees than PD1 blockade. In addition, TIM3 blockade led to upregulation of CD28, which is critical for the rescue of Tex through costimulation, while PD1 blockade did not.
We then analyzed the clinical outcomes of all 34 MZL patients. The frequency of PD1-TIM3+ cells was lower among observed patients who failed to achieve an event-free survival (EFS) of 12 months (EFS12) (0.8% vs 1.7%, p=0.02) and among treated patients that failed an EFS of 24 months (EFS24) (0.9 vs 1.7%, p=0.03), when compared to patients that reached these endpoints. Higher levels of PD1-TIM3+ cells were also associated with a trend towards improved overall survival (OS). Five-year OS was 88% vs. 69% in patients with PD1-TIM3+ levels >2% and <2% of total CD8 cells respectively (p=0.1) (Fig 3).
Our study provides insights into the mechanisms of failure of PD1i in MZL and identifies a novel population of Tex with high reactivation potential which is clinically relevant and could serve as a target for future immunotherapies in iNHL.
Anagnostou:American Society of Hematology, Mayo Clinic/Iowa Lymphoma SPORE, Mayo Clinic Immune Monitoring Core, Mayo Clinic Hematology Small Grant: Research Funding. Novak:Celgene Coorperation: Research Funding. Ansell:Mayo Clinic Rochester: Employment; Trillium: Research Funding; Seattle Genetics: Research Funding; Regeneron: Research Funding; Bristol-Myers Squibb: Research Funding; Seattle Genetics: Research Funding; Mayo Clinic Rochester: Employment; Affimed: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; LAM Therapeutics: Research Funding; Mayo Clinic Rochester: Employment; Bristol-Myers Squibb: Research Funding; Mayo Clinic Rochester: Employment; Trillium: Research Funding; Seattle Genetics: Research Funding; Bristol-Myers Squibb: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Regeneron: Research Funding; Mayo Clinic Rochester: Employment; Seattle Genetics: Research Funding; Regeneron: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; Trillium: Research Funding; Mayo Clinic Rochester: Employment; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; Trillium: Research Funding; Regeneron: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; Mayo Clinic Rochester: Employment; Bristol-Myers Squibb: Research Funding; Bristol-Myers Squibb: Research Funding; Regeneron: Research Funding; Mayo Clinic Rochester: Employment; Regeneron: Research Funding; Trillium: Research Funding; Seattle Genetics: Research Funding; Seattle Genetics: Research Funding; Trillium: Research Funding; Regeneron: Research Funding; Affimed: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; LAM Therapeutics: Research Funding; Regeneron: Research Funding; Affimed: Research Funding; Trillium: Research Funding; Bristol-Myers Squibb: Research Funding; LAM Therapeutics: Research Funding; Affimed: Research Funding; Affimed: Research Funding; Seattle Genetics: Research Funding; LAM Therapeutics: Research Funding; Bristol-Myers Squibb: Research Funding; Trillium: Research Funding; LAM Therapeutics: Research Funding; Bristol-Myers Squibb: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.