Background
Treatment decisions in Multiple Myeloma (MM) have been driven by a patient's age or ability to tolerate therapy, but as yet, not by their tumour genetics, even though understanding of the prognostic utility of genetic features continues to develop. The ability to identify genetic prognostic markers and potential therapeutic targets in a timely fashion within a health service is a vital step in delivering precision medicine to patients. The aims of this study were to assess the implementation of clinical grade whole genome sequencing (WGS) data at diagnosis to replicate, and ultimately replace, standard-of-care Fluorescence In Situ Hybridisation (FISH) cytogenetics; to provide markers for prognostication or MRD; and to identify actionable targets for clinical trials in precision therapy of MM.
Methods
Bone marrow aspirates from patients with newly-diagnosed myeloma were collected between June 2017 and April 2018 in a single tertiary hospital in the United Kingdom. The population comprised seven male and seven female patients with a mean age of 78 years. From the first-draw of bone marrow aspirate, standard-of-care FISH cytogenetic analyses were performed locally according to criteria from the International Myeloma Working Group (IMWG). From the remnant aspirate samples, CD138-positive plasma cells were enriched by magnetic bead-sorting and genomic DNA was extracted locally for WGS with a success rate of approximately 70%.
Fourteen samples underwent successful plasma cell purification (78 - 99% morphological purity) with a yield of at least 0.5 μg DNA. To identify germline genomic variants, DNA was extracted from peripheral blood samples obtained simultaneously. WGS was performed at a centralised facility and mean coverage for germline samples was 35.1x and for plasma cell-enriched samples was 100.9x. Conventional cytogenetic FISH data were compared with genomic data for chromosome-level alterations. Identified somatic variants were automatically cross-referenced against publicly available databases that describe somatic mutations in cancer as well as a virtual panel of potentially actionable therapeutic targets including : NRAS, KRAS, BRAF, CDKN2C, FGFR3 and IDH2.
Results
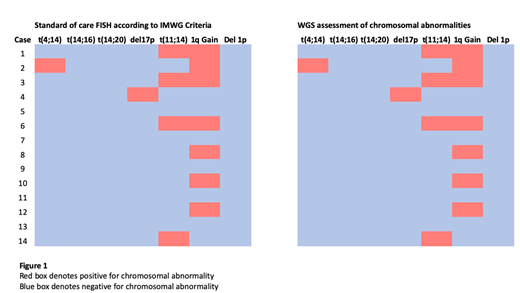
In paired samples, WGS replicated all 13 translocation and chromosomal loss/gain events identified by FISH (Figure 1). Furthermore, three translocations involving the IGH locus suggested by FISH analysis were characterised by WGS. Using samples derived from surplus material, fast-track turnaround of 14 days was attainable.
Five patients had no identifiable marker by FISH cytogenetics. In these patients, WGS found all five to possess somatic variants could be used as prognostic or potential Measurable Residual Disease (MRD) markers.
Nine patients exhibited somatic variants in genes that may be subject to targetable therapy as determined by trials available on ClinicalTrials.gov: five NRAS, two KRAS, one BRAF and one FGFR3 mutation as of July 2019.
Conclusion
WGS assessment of newly diagnosed myeloma provides accurate, timely and actionable information beyond what is available from standard-of-care FISH.
The application of WGS to myeloma diagnostics presents a number of advantages. Firstly, WGS can replicate and exceed existing myeloma FISH assessment of translocation and loss/gain events in a prompt turnaround time. Secondly, germline variants are deducted from tumour variants to provide a gold standard description of the somatic mutational landscape in the patient's disease. Thirdly, virtual panels of known somatic variants can be applied to the data as knowledge accumulates about the role of specific mutations on prognosis or therapeutic response.
We demonstrate that centrally provided WGS and its analysis can be incorporated into routine local assessment of newly diagnosed myeloma. Therefore, these observations show that such technology has the potential to be rapidly scalable across existing hospital networks.
Gooding:Celgene Corporation: Research Funding. Ramasamy:Celgene: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Research Grants; Janssen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Research Grants; Amgen: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Research Grants; Takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Research Grants; Abbvie: Honoraria, Membership on an entity's Board of Directors or advisory committees; Oncopeptides: Honoraria, Membership on an entity's Board of Directors or advisory committees; Sanofi: Honoraria, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.