Introduction: Myelodysplastic syndromes (MDS) are a heterogenous group of blood disorders defined by peripheral cytopenia(s), bone marrow failure, morphological dysplasia, and risk of progression. To understand the genetic, epigenetic and biological factors associated with the initiation and progression of MDS, NHLBI created the National MDS Study (NCT02775383). This is a prospective cohort study conducted at 92 community hospitals and 29 academic centers enrolling patients undergoing diagnostic work up for suspected MDS or MDS/myeloproliferative neoplasm (MPN) overlap syndrome. Eligible patients have yet to receive any therapy directed at their cytopenias. Previously untreated cytopenic participants underwent centralized histopathology and data review at the time of enrollment for assignment into distinct subcategories: MDS, MDS/MPN overlap, AML, and Other. Targeted exon sequencing of 96 genes was performed using marrow specimens from the first 300 consecutive individuals in the study. Here we report the genetic mutations for this cohort.
Methods: NovaSeq 6000 was used for deep sequencing at a mean coverage of 1,286X and mean breadth (bases covered at ≥100X) of 99.8%. Reads were aligned against build GRCh38 using BWA-MEM, and VarScan2 was used to detect SNVs and INDELS. Variants were filtered for those with an allele base quality of >25 in combination with rule-based and manual review criteria. Subjects in the Other category without an identified malignancy were considered clonal cytopenias of undetermined significance (CCUS) when a mutation or a clonal cytogenetic change was present. Fisher's exact and Wilcoxon rank sum tests in combination with Bonferroni correction were applied to compare groups.
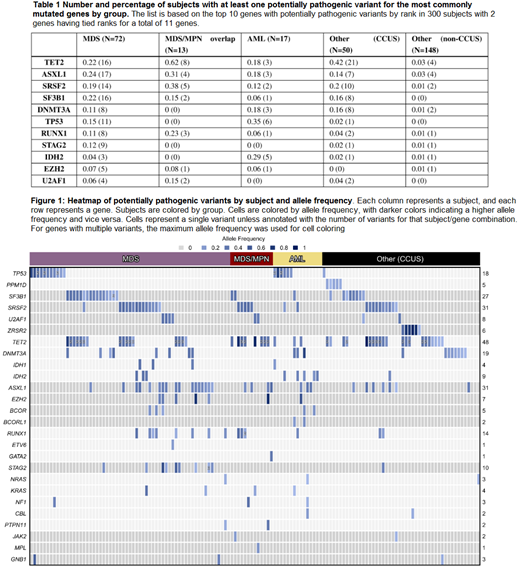
Results: A total of 350 putative nonsynonymous pathogenic variants in 36 genes with an allele frequency of >.05 were identified across 150 patients (50%). At least one variant was noted in the following proportion of individuals: 61/72 (85%) with MDS, 13/13 (100%) with MDS/MPN, 15/17 (88%) with AML, and 61/198 (31%) in the Other category, of which 48 were CCUS and 13 were other cancers. Two CCUS patients only had a cytogenetic abnormality. Table 1 shows the distribution of variants in each subcategory of patients for the most commonly mutated genes in our cohort of 300 subjects. Mutations in these genes were enriched in specific groups: SF3B1, STAG2,TP53, and ASXL1 in MDS; TET2 in MDS/MPN; and IDH2 and TP53 in AML (one-sided p<0.0012). None were enriched in the Other group. Within the CCUS subset, 21 genes were mutated, with 37 of 50 (74%) patients having a mutation in TET2, ASXL1, SRSF2, SF3B1, or DNMT3A. The heatmap presented in Figure 1 summarizes variants by subject and allele frequency.
Pair-wise comparisons of baseline characteristics of subjects between MDS, MDS/MPN, AML, or CCUS groups revealed no significant differences for age or sex. The CCUS group had significantly higher hemoglobins than the MDS group with median hemoglobin levels of 11.35 and 9.40 g/dL, respectively (p<0.0084). AML had significantly lower platelets compared to all other groups (median of 53 vs. > 110x109/L , respectively, p<0.0084). All groups significantly differed in their median ANC with AML having the lowest (0.8x109/L), followed by MDS (1.5x109/L), CCUS (2.45x109/L), and MDS/MPN overlap (5.95x109/L) (p<0.0084).
There was no difference in the median number of variants per patient between groups or correlation with age (rs=0.11, p=0.18). The maximum variant allele frequency (maxVAF) per patient was highest in the MDS/MPN group (median = 0.42, range = 0.38-0.91) and lowest in the CCUS group (median = 0.37, range =0.06-0.98) with the MDS/MPN group having a significantly higher maxVAF compared to the MDS and the CCUS groups (p<0.0084). The proportions of subjects with mutations was similar for those who had abnormal (92% [34/37]) and normal (91% [80/88]) cytogenetics.
Conclusions: Incorporation of gene-panel sequencing in the comprehensive evaluation of 300 adult cytopenic patients identified half of the cohort with potentially pathogenic variants. Ultimately, a diagnosis of CCUS was possible in 48 of 183 subjects (26%) not diagnosed with MDS, MDS/MPN overlap syndrome, AML, other cancers or clonal cytogenetics. This study continues to serially bank samples from patients with CCUS, in addition to MDS, MDS/MPN, and ICUS, with the goal to better understand the natural history of these diseases and their progression.
Lindsley:Jazz Pharmaceuticals: Research Funding; Takeda Pharmaceuticals: Consultancy; Medlmmune: Research Funding. Bejar:Celgene: Consultancy; Takeda Pharmaceuticals: Research Funding; AbbVie/Genentech: Consultancy, Honoraria; Astex/Otsuka: Consultancy; Modus Outcomes: Consultancy; Daiichi-Sankyo: Consultancy. Al Baghdadi:Celgene: Consultancy, Honoraria; Heron: Consultancy, Honoraria; Tracon: Equity Ownership; Epizyme: Equity Ownership; Bristol Myer Squibb: Consultancy, Honoraria; Sunesis: Equity Ownership; Portola: Equity Ownership; Heron therapeutics: Equity Ownership; Cardinal health: Consultancy, Honoraria; Bristol Myer Squibb: Equity Ownership; Celgene: Equity Ownership; Spectrum pharmaceutical: Equity Ownership; Astrazeneca: Equity Ownership; Seattle genetics: Equity Ownership; Roche: Consultancy, Honoraria. DeZern:Astex Pharmaceuticals, Inc.: Consultancy; Celgene: Consultancy. Foran:Agios: Honoraria, Research Funding. Gore:Celgene Corporation: Consultancy, Research Funding. Komrokji:DSI: Consultancy; Agios: Consultancy; Novartis: Speakers Bureau; JAZZ: Speakers Bureau; pfizer: Consultancy; celgene: Consultancy; Incyte: Consultancy; JAZZ: Consultancy. Maciejewski:Alexion: Consultancy; Novartis: Consultancy. Starczynowski:Kurome Therapeutics: Consultancy. Sekeres:Millenium: Membership on an entity's Board of Directors or advisory committees; Syros: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.