Introduction
Translocation of intestinal bacteria across impaired mucosal barriers has long been believed to occur following exposure to chemotherapy. Consistent with this, we and others have previously reported that expansions of potentially pathogenic bacteria within the gastrointestinal microbiome precedes bloodstream infection. While the blood microbiome represents a rich area for investigation, detailed unbiased characterization of the blood microbiome has not previously been possible. Here, we sought to investigate the relationship between the blood and stool microbiome in patients undergoing allogeneic hematopoietic stem cell transplantation (HCT) who are at high risk for gut barrier dysfunction, severe infection, and the immunological complications of transplantation including graft-versus-host disease (GVHD). We show preliminary data suggesting that the blood microbiome may hold biomarkers for gut integrity. Subsequently, it may offer microbiological data supporting causative organisms in "culture-negative" fevers, or in itself, perform an immunomodulatory role acting as a damage/pathogen associated molecular pattern (DAMP/PAMP).
Methods
We sequenced serially-collected plasma and stool samples (n = 61 unique samples of each type) from a cohort of 19 patients who underwent allogeneic HCT at our center. Microbial cell-free DNA (mcfDNA) was extracted from plasma and sequenced using a next generation sequencing assay (Karius, Inc, Redwood City, CA). After sequences are processed, mcfDNA abundances are reported in molecules per microliter. Stool samples underwent were profiled using 16S-targeted sequencing (V4-V5 region) on the Illumina MISEQ platform and analyzed using the DADA2 pipeline. Association of blood microbial burden with clinical factors was assessed using a Wilcoxon rank sum test.
Results
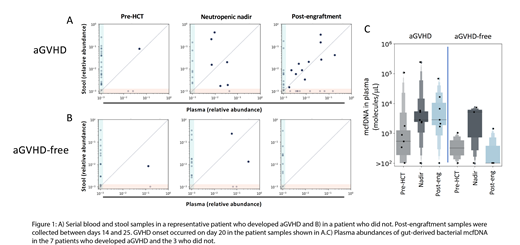
We confirmed a high sequence homology between the DNA found in plasma and stool samples, thus demonstrating that circulating mcfDNA is gut-derived in these patients. By comparing microbial sequences from paired plasma and stool samples collected equivalent time points during the neutropenic nadir after HCT, we observed a correlation between the abundance of bacterial mcfDNA in plasma and DNA from the same microbes in stool. Of note, this occurred independently of conditioning regimen intensity, and was observed in patients who had received myeloablative, non-myeloablative and reduced intensity therapy. We assessed the association of bacterial mcfDNA burden (only considering sequences common to stool sequences) with various clinical factors. Translocation was significantly higher in patients who experienced pre-engraftment mucositis (any observed grade) compared with those who did not (p = 0.02, n = 5 in the mucositis group, any grade, and 12 in the mucositis-free group), but there was no relationship between the degree of translocation and HCT-CI, conditioning intensity, donor type, age or pre-engraftment fevers. We next asked whether translocation events were associated with acute GVHD (aGVHD) by assessing a subgroup of patients who received unmodified grafts (peripheral blood stem cell or bone marrow; n = 10) and tracked the degree of translocation from the gut to the blood stream pre-transplant, during the neutropenic nadir, and following engraftment. As shown in Figure 1, we observed low pre-transplant translocation (as quantified by the total bacterial DNA abundance in plasma where sequences were also shared in stool samples), and a marked increase during neutropenic nadir. Remarkably, even in this small cohort, we observe a higher burden of translocation in the post-engraftment period in patients who subsequently develop aGVHD, while it decreased to baseline levels in those who do not (n = 7 in the aGVHD group; n = 3 aGVHD-free; p = 0.05).
Conclusions
Here we demonstrate the first use of a culture-free molecular assay to track the blood microbiome and identify features of the circulating mcfDNA that correlate with the microbial DNA in stool samples. Despite the small sample size, these data suggest that the maintenance of a high mcfDNA-burden beyond the neutropenic nadir is associated with subsequent GVHD development, and provides some evidence for early gut barrier dysfunction that permits translocation in these patients.
Blair:Karius, Inc: Employment. Peled:Seres Therapeutics: Other: IP licensing fees, Research Funding. Giardina:Seres Therapeutics: Other: Salary funding. Slingerland:Seres Therapeutics: Other: Salary supported by Seres funding. Hollemon:Karius, Inc: Employment. Ho:Karius, Inc: Employment. Bercovici:Karius, Inc: Employment. Ahmed:Karius, Inc: Employment. Hong:Karius, Inc: Employment. Giralt:Celgene: Consultancy, Research Funding; Takeda: Consultancy; Sanofi: Consultancy, Research Funding; Amgen: Consultancy, Research Funding. van den Brink:Therakos: Consultancy, Honoraria; Merck & Co, Inc.: Consultancy, Honoraria; Amgen: Consultancy, Honoraria; Acute Leukemia Forum (ALF): Consultancy, Honoraria; Magenta and DKMS Medical Council: Membership on an entity's Board of Directors or advisory committees; Novartis: Consultancy, Honoraria; Seres Therapeutics: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Flagship Ventures: Consultancy, Honoraria; Juno Therapeutics: Other: Licensing; Evelo: Consultancy, Honoraria; Jazz Pharmaceuticals: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.