Background: Effective treatment of relapsed/refractory (r/r) DLBCL remains a major unmet need. Checkpoint blockade therapy (CBT) leads to durable responses in a small subset of r/r DLBCL patients, but limited understanding of predictive biomarkers and characteristics of host immune responses have slowed development of DLBCL immunotherapy. We previously described a subset of "T-cell inflamed" DLBCLs marked by PD-L1 gene alterations and increased likelihood of response to CBT. In this study, we aimed to identify and group gene expression patterns associated with immune features in a large number of DLBCL cases available in published datasets. We investigate the immunogenomic features of each group and corroborate findings in primary DLBCLs.
Methods: Gene sets reflecting a broad array of activation states or subtypes of tumor-infiltrating immune cells were selected from previous studies (n = 143). Expression by case was scored for each set by applying gene set variation analysis (GSVA) to previously published DLBCL bulk RNAseq profiles (n = 1189). The resulting score matrix was reduced with principal component analysis (PCA); the first 10 components were used to hierarchically cluster each case into related groups. Immune cell fractions were estimated from RNAseq counts via deconvolution analysis. Differentially expressed genes (DEG) for each cluster were identified by false discovery rate (FDR) < 0.05 and log2 fold change of > 1.5. The cytolytic gene expression (CYT) score, associated with T-cell immunity against solid tumors, was computed for each case. Protein-coding mutations found in ≥ 5% of cases by whole exome sequencing analysis were filtered for driver mutations (MutSig2CV q-value < 0.1). PD-L1 gene amplification status was determined where copy number array data were available (n = 471). RNAseq (Illumina HiSeq 2000 platform) was performed on 24 fixed and embedded treatment-naive DLBCL tumors from an institutional biorepository. GSVA scores were projected onto the PCA and clusters assigned. CD4+ and CD8+ T-cells per high-power field (HPF) were assessed by immunohistochemistry (IHC).
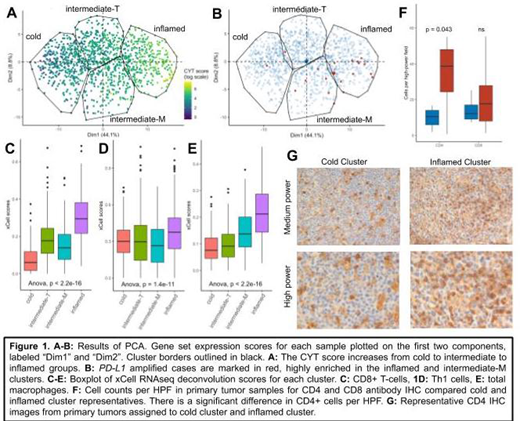
Results: Four clusters of immune gene set expression were identified in existing DLBCL datasets, termed "inflamed", "intermediate-M", "intermediate-T" and "cold". There is no association of any cluster with a difference in overall survival or enrichment in a cell of origin subtype. The inflamed cluster has the highest mean CYT score (Fig 1A, p < 0.001) and highest deconvolution-estimated fraction of CD8+ T-cells, M1 and M2 macrophages, dendritic cells, T-helper 1, and T-helper 2 cells (Fig 1C-E, p < 0.001). Significantly upregulated DEGs include CXCL9, CXCL10, CCL8, and CXCR6, which have been associated with a T-cell inflamed phenotype in solid tumors. Immune escape mechanisms in the inflamed cluster are suggested by upregulated DEGs of VSIG4 and IDO1, and significant enrichment for PD-L1 gene amplifications compared to the cold cluster (Fig 1B, 8.6% vs 1%, p = 0.01). The cold cluster has the lowest mean CYT score (p < 0.001), and lowest deconvolution scores for CD8+ T-cells, M1 and M2 macrophages, and dendritic cells (p < 0.001). The cold cluster harbors more mutations in MYD88 (27%),TMSB4X (11.6%), and FOXO1 (8%) and fewer SOCS1 (7%) mutations than other clusters (FDR < 0.25). Intermediate-T and intermediate-M clusters share mid-range values of estimated CD8+ T cells and CYT scores, but intermediate-M contained more frequent PD-L1 amplifications (Fig 1B, 9% vs 0.9%, p = 0.02), lower estimated Th1 fraction (p < 0.001), and higher estimated total macrophages (p < 0.001) than intermediate-T. Representatives of each cluster were identified in primary DLBCL tumors (n = 24). Mean CD4+ T-cell count per HPF was higher in inflamed cluster DLBCLs compared to cold (45.1 vs 10.1, p = 0.032). A non-significant increase in CD8+ cells was also seen (21.3 vs 14.2 per HPF).
Conclusion: In this first comprehensive immunogenomic study of DLBCL, we define differences in host immune response by gene set expression, associate oncogenic mutations with immune exclusion, and discover expression of a number of immune escape genes in inflamed cases. Primary samples analyzed to date support the immune response patterns found by computational analysis. A greater understanding of heterogeneity in host response to DLBCL may help identify subsets of DLBCLs with inherent vulnerability to CBT and other immunotherapies.
Smith:Portola Pharmaceuticals: Research Funding. Kline:Merck: Honoraria; Merck: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.