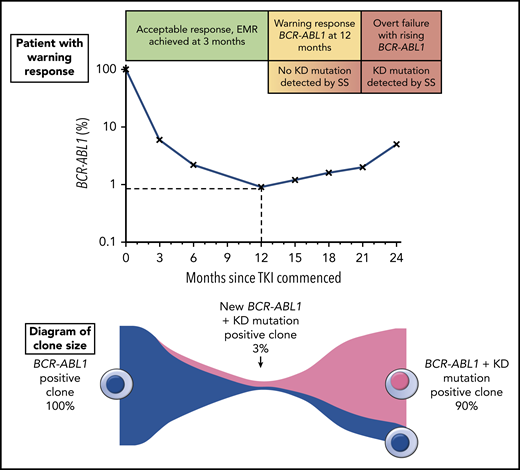
In this issue of Blood, Soverini et al illustrate the potential utility of next-generation sequencing (NGS)-based BCR-ABL1 kinase domain (KD) mutation analysis of samples from patients with chronic myeloid leukemia (CML) experiencing either treatment failure (F) or warning (W) responses.1
The top portion of the figure illustrates a hypothetical patient diagnosed with chronic phase CML and commenced on TKI therapy. The patient had a warning response to TKI, based on the 2013 European LeukemiaNet criteria due to of failure to achieve a major molecular response9 (BCR-ABL1 ≤0.1%) by 12 months. Although the patient’s response plateaued for several months, there was a subsequent progressive increase in BCR-ABL1 consistent with overt treatment failure. The lower portion of the figure illustrates the clone sizes at various stages of therapy. At the time of the warning response, a KD mutation emerged with an allele frequency of ∼3%, but because this was not detectable by SS, the same therapy was continued until the resistant clone expanded enough to be eventually detectable by SS. Earlier intervention would likely have prevented overt treatment failure by initiating an appropriate therapy switch when the new low-level KD mutation was detected.
The top portion of the figure illustrates a hypothetical patient diagnosed with chronic phase CML and commenced on TKI therapy. The patient had a warning response to TKI, based on the 2013 European LeukemiaNet criteria due to of failure to achieve a major molecular response9 (BCR-ABL1 ≤0.1%) by 12 months. Although the patient’s response plateaued for several months, there was a subsequent progressive increase in BCR-ABL1 consistent with overt treatment failure. The lower portion of the figure illustrates the clone sizes at various stages of therapy. At the time of the warning response, a KD mutation emerged with an allele frequency of ∼3%, but because this was not detectable by SS, the same therapy was continued until the resistant clone expanded enough to be eventually detectable by SS. Earlier intervention would likely have prevented overt treatment failure by initiating an appropriate therapy switch when the new low-level KD mutation was detected.
Patients with resistance to treatment or disease progression do not always have an overt resistance mechanism that has been identified. KD mutations, the best understood mechanism of resistance in CML, are identified in only ∼50% of patients for who imatinib treatment has failed,2 although this may be an underestimate because of the limited sensitivity of Sanger sequencing (SS). The resistance profile of specific KD mutations can be used to select subsequent tyrosine kinase inhibitor (TKI) therapy because resistant clones can be expected to expand if an inappropriate TKI is continued or selected for next-line therapy (see figure). A more sensitive mass spectrometry assay for KD mutations was developed that lowered the detection limit to 0.2% for the majority of mutations,3 and was highly clinically predictive, but not widely available.
The NEXT-in-CML study led by Soverini et al was a prospective assessment of NGS compared with SS (the current gold standard method) in identifying KD mutations in CML patients with molecular responses to TKI therapy who were classified as either W or F.3 The first phase of the study involved viability assessments of the NGS-based mutation analysis across collaborating laboratories, which confirmed the accuracy and reproducibility of the assay. Although SS was able to identify KD mutations in 25% of patients in the clinical phase of the study, NGS detected low-level KD mutations in an additional 22%. NGS also identified additional low-level mutations in 48% of the patients already found to have KD mutations on SS. Moreover, patients classified as F were more likely to harbor multiple mutations and had a higher risk of experiencing subsequent treatment failure, which supports previous observations.4 Longitudinal assessments of the kinetics of low-level mutations demonstrated persistence and expansion of the mutation known to be resistant to the selected TKI, as predicted.
This comprehensive prospective study provides an alternative method for sensitive KD mutation testing, one that is perhaps easier to adopt given the number of institutions that have established NGS testing for other purposes. Earlier detection of low-level KD mutations will inform clinicians of the need to alter the current therapy before the disease progresses to blast crisis CML, which is a real possibility as shown in this study. The dismal outcomes associated with blast crisis CML5 may be circumvented by an earlier switch to an optimal TKI, which would reduce transformation rates and improve survival.
However, several limitations associated with NGS make routine application of this technology for the proposed purpose challenging. Although Soverini et al were able to provide results within an average of 11 days (range, 7-24 days), many laboratories would struggle to achieve this turnaround time,6 which would thwart the feasibility of obtaining an actionable result before overt treatment failure or disease progression. Moreover, the costs associated with NGS can be prohibitive in a diagnostic setting; thus, batch testing would be required to reduce the expense associated with testing each sample separately, adding to the delay in obtaining results. In comparison, SS is fast and inexpensive, although it has a mutation detection limit of 10% to 20%,7,8 which negates its applicability in low-level mutation testing. Before NGS-based assays can be adopted into mainstream diagnostics, these issues will need to be addressed by each institution that intends to implement them for the purpose of KD mutation testing.
An issue that now needs to be addressed is the actual consensus definition of low-level mutation. The NGS-based assay used by Soverini et al in their study, classified low-level KD mutations as being undetectable by SS, which correlates to a variant allele frequency of 3% to 20%. However, as technology improves, it is likely that the sensitivity of NGS will increase, with the potential to detect mutations that have an allele frequency considerably less than 3%. What should the actionable threshold for recommending a switch in TKIs? Furthermore, should we be screening patients deemed to be at high risk for treatment failure at earlier time points, even before a warning response is recognized? This could conceivably include screening selected patients at diagnosis to detect low-level actionable KD mutations that would optimize TKI selection and prevent the development of TKI resistance. Although these issues will need to be considered in the near future, the findings from Soverini et al make a strong argument to support the use of NGS in patients who do not achieve optimal molecular responses; facilitating rational selection of TKI and ideally prevent overt drug resistance and disease progression.
Conflict-of-interest disclosure: N.S. received honoraria from Novartis and travel and accommodation expenses from Novartis, Gilead, Amgen, and Janssen. T.P.H. has served as a consultant for and has received research funding and honoraria from Novartis and Bristol-Myers Squibb.