Abstract
Introduction: Drug resistance at relapse is a major cause of mortality in AML. Previous genomic profiling of AML patient samples revealed that in many cases the mutational profile did not change between diagnosis and relapse (Parkin et al., Blood, 2013, Nuno K et al., Blood, 2020), indicating that epigenetic changes can have a substantial contribution to acquired drug resistance and refractory disease at relapse. In order to uncover such functional alterations, longitudinal AML patient samples collected at diagnosis, during remission and relapse were analysed with single cell transcriptomics (scRANseq) in order to identify how the molecular wiring of AML cells evolve during disease progression and what alterations make them drug resistant.
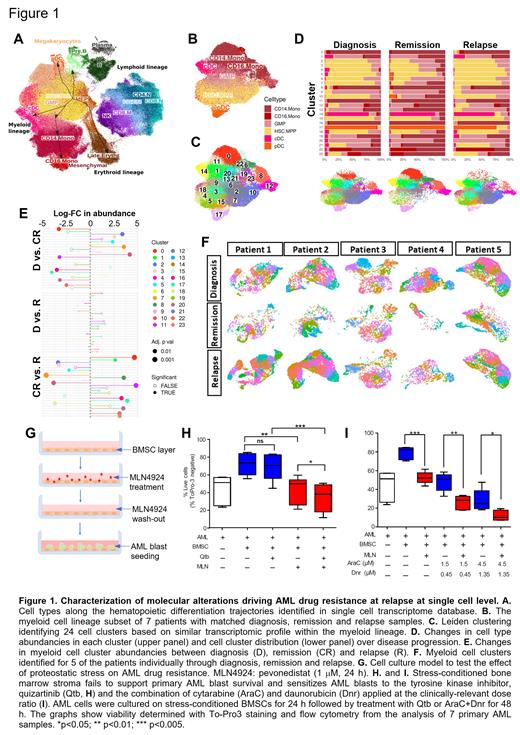
Methods: scRNAseq was carried out (10x Genomics) on mononuclear cell fractions from bone marrow aspirates of 7 AML patients (from the Finnish Hematology Registry and Clinical Biobank) who received AraC-based induction therapy, achieved CR but later relapsed. Raw data was QC processed (CellRanger), batch corrected (scArches, Harmony). Cell types were identified as described by Granja et al., Nat. Biotechn., 2020 (Fig 1A), the myeloid cell lineage was selected, clusters identified (Leiden) and differentially expressed genes identified (EdgeR). For mechanistic studies primary AML blasts were cultured with bone marrow stromal cells (BMSC) in a system able to recapitulate in vivo chemotherapy resistance (Dhami et al., Br. J, Haematol, 2020).
Results: The scRNAseq analysis generated transcriptome for over 67,000 myeloid-lineage cells (hematopoietic/leukemic stem cells (HSC/LSC), multipotent progenitors (MPP), granulocyte-monocyte progenitors (GMP), monocytes (CD14, CD16 subsets), conventional and plasmacytoid dendritic cells; c/pDCs) including 21K, 26K and 22K cells at diagnosis, remission and relapse, respectively (Fig 1B). To identify drug resistant AML clones, we integrated the myeloid lineage cells from the 7 patients and identified clusters (Fig 1C). Of the 24 clusters found, the ones rich in differentiated cells became abundant at remission, while clusters rich in low-differentiation status cells (HSC/LSC, MPP, GMP) diminished. Upon relapse however this trend reversed and all main clusters present at diagnosis returned, indicating that cells from all clusters survived chemotherapy or drug-resistant LSCs could reinstate the full disease spectrum (Fig 1D, 1E). Analysis of AML cell clusters in each patient individually confirmed this pattern with nearly all clusters returning at relapse, excluding the possibility that integration masked patient-level heterogeneity/patient-specific cluster patterns (Fig 1F).
Despite the identical cluster distribution between diagnosis and relapse, all 7 patients showed refractory disease at relapse, indicating that small scale molecular alterations, which are not substantial enough to segregate the cells into a new cluster are sufficient to drive drug resistance. To identify these changes, genes differentially expressed and associated gene ontology (GO) terms between diagnosis and relapse were identified for each cluster. The analysis found GO terms commonly regulated in multiple clusters, most notably myeloid cell activation, mitochondrial ATP metabolism, cellular respiration, mRNA maturation and translation.
In order to determine whether targeting the protein synthesis machinery, as a process linked to integrated stress response and HSC maintenance, can be exploited to eliminate drug resistant AML cells, we exposed BMSCs to transient proteostatic stress induced by the NEDDylation inhibitor pevonedistat (1 mM for 24 h), cultured primary AML blasts on the stress-conditioned BMSCs and exposed them to drug treatments (Fig 1G). Stress-conditioned BMSCs lost their ability to support AML blast survival and to protect them from tyrosine kinase inhibitors (quizartinib) and the combination of AraC+daunorubicin.
Conclusions: We found that AML cell clusters that already exist at diagnosis re-emerge at relapse, although with re-occurring molecular alterations that can provide drug resistance resulting in refractory disease. Targeting one of these pathways, proteostasis, could break the protective interaction between BMSCs and AML blasts resulting in reduced AML survival and enhanced drug sensitivity, representing a potential actionable vulnerability of drug resistant AML cells.
Szegezdi: Bristol Myers Squibb: Research Funding; ONK Therapeutics: Research Funding.