Abstract
Background: Approximately 20% of AML patients do not respond to induction chemotherapy (primary resistance) and 40-60% of patients develop secondary resistance, eventually leading to relapse followed by refractory disease (RR-AML). Diversified molecular mechanisms have been proposed for drug resistance and RR phenotype. However, we still cannot predict when relapse will occur, nor which patients will become resistant to therapy.
Single-cell multi-omic (ScMo) profiling may provide new insights into our understanding of hematopoietic stem cell (HSC) differentiation trajectories, tumor heterogeneity and clonal evolution. Here we applied ScMo to profile bone marrow (BM) from AML patients and healthy controls.
Methods: AML samples were collected at diagnosis with institutional IRB approval. Cells were stained with a panel of 62 DNA barcoded antibodies and 10x Genomics Single Cell 3' Library Kit v3 was used to generate ScMo data. After normalization, clusters were identified using Uniform Manifold Approximation and Projection (UMAP) and annotated using MapCell (Koh and Hoon, 2019). We analyzed 23,933 cells from 4 adult AML BM samples, and 39,522 cells from 2 healthy adults and 3 sorted CD34+ normal BM samples. Gene set enrichment analysis (GSEA) and Enrichr program were used to examine underlying pathways among differentially expressed genes between healthy and AML samples.
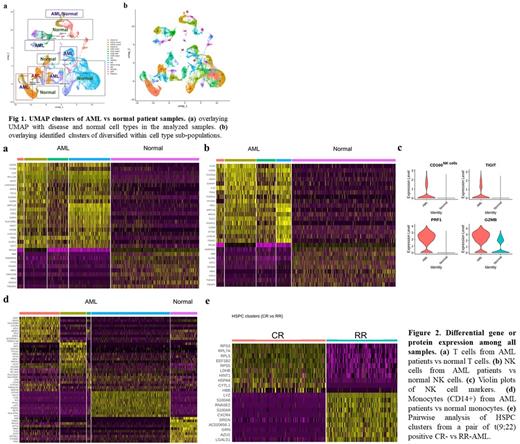
Results: We identified 16 cell types between the AML and normal samples (Fig 1a) amongst 45 clusters in the UMAP projection (Fig 1b). Comparative analysis of the T cell clusters in AML samples with healthy BM cells identified an "AML T-cell signature" with over-expression of genes such as granzymes, NK/T cell markers, chemokine and cytokine, proteinase and proteinase inhibitor (Fig 2a). Among them, IL32 is known to be involved in activation-induced cell death in T cells and has immunosuppressive role, while CD8+ GZMB+ and CD8+ GZMK+ cells are considered as dysfunctional or pre-dysfunctional T cells. Indeed, Enrichr analysis showed the top rank of phenotype term - "decreased cytotoxic T cell cytolysis".
We next examined whether NK cells, are similarly dysfunctional in the AML ecosystem. The "AML NK cell signature" includes Fc Fragment family, IFN-stimulated genes (ISGs), the effector protein-encoding genes and other genes when compared to normal NK cells (Fig 2b). GSEA analysis revealed "PD-1 signalling" among the top 5 ranked pathways in AML-NK cells, though no increase in PD-1 protein nor PDCD1 gene were identified in these cells. Inhibitory receptor CD160 was expressed higher in AML samples along with exhaustion (dysfunction) associated genes TIGIT, PRF1 and GZMB (Fig 2c). Enrichr analysis uncovered enrichment of "abnormal NK cell physiology and "impaired natural killer cell mediated cytotoxicity". Similarly, the "AML monocyte signature" was significantly enriched with genes in "Tumor Infiltrating Macrophages in Cancer Progression and Immune Escape" and "Myeloid Derived Suppressor Cells in Cancer Immune Escape".
We also analyzed HSPC component in one pair of cytogenetically matched, untreated complete remission (CR) /RR AML pair (Fig 2d). Notably, half of the 10 genes overexpressed in RR-AML, CXCR4, LGALS1, S100A8, S100A9, SRGN (Serglycin), regulate cell-matrix interaction and play pivotal roles in leukemic cells homing bone marrow niche. The first 4 of these genes have been demonstrated as prognostic indicators of poor survival and associated with chemo-resistance and anti-apoptotic function. Furthermore, single-cell trajectory analysis of this CR/RR pair illustrated a change in differentiation pattern of HSPCs in CR-AML to monocytes in RR-AML. We are currently analyzing more AML samples to validate these findings.
Conclusions: Our ScMo analysis demonstrates that the immune cells are systematically reprogrammed and functionally comprised in the AML ecosystem. Upregulation of BM niche factors could be the underlying mechanism for RR-AML. Thus, reversing the inhibited immune system is an important strategy for AML therapy and targeting leukemic cell-BM niche interaction should be considered for cases with high expression of these molecules on AML HSPCs.
Note: J.Z. and J.A.S. share co-first authorship.
Scolnick: Proteona Pte Ltd: Current holder of individual stocks in a privately-held company. Xu: Proteona Pte Ltd: Current Employment. Ooi: Jansen: Honoraria; Teva Pharmaceuticals: Honoraria; GSK: Honoraria; Abbvie: Honoraria; Amgen: Honoraria. Lovci: Proteona Pte Ltd: Current Employment. Chng: Aslan: Research Funding; Takeda: Honoraria; Johnson & Johnson: Honoraria, Research Funding; BMS/Celgene: Honoraria, Research Funding; Amgen: Honoraria; Novartis: Honoraria, Research Funding; Antengene: Honoraria; Pfizer: Honoraria; Sanofi: Honoraria; AbbVie: Honoraria.