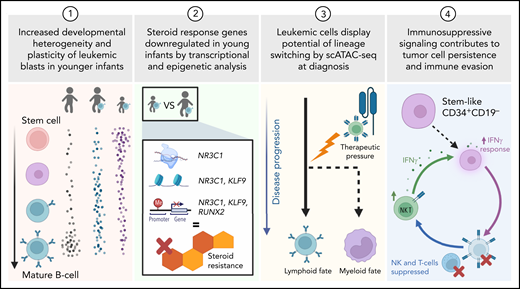
In this issue of Blood, Chen et al use multiomic single-cell analysis to characterize KMT2A-rearranged (KMT2A-r) acute lymphoblastic leukemia (ALL) and show that plasticity, steroid resistance, and immunosuppressive stem-like cells are highest in the youngest patients.1
Leukemia in the pediatric population differs from that in the adult population in both incidence and prognosis. Even within the pediatric population, large differences in both factors have been identified between infants (<1 year) and older children (≥1 year). KMT2A-r leukemias make up ∼70% of cases of infant acute leukemias but only 10% of cases in older children.2 Event-free survival is >90% for older children with ALL but 36% in infants with KMT2A-r ALL and <20% for patients younger than 3 months.3,4 Despite the evaluation of new treatment strategies to improve prognosis, including chemotherapeutics and targeted therapies, the outcome remains poor for infants with ALL. There is a critical need for further research regarding factors underlying age-related differences in KMT2A-r ALL that may guide improved treatments for the youngest patients.
In this study, Chen et al apply multiomic profiling to gather cell-level information on gene expression, chromatin accessibility, DNA methylation, and transcript fusions in patients with KMT2A-r ALL. Parallel measurements of these features allow researchers to form systems-level visions of how each cell works individually and with its neighbors, progenitors, and progeny. The authors use these data to uncover four key factors that are correlated with patient age at diagnosis and may determine outcome (see figure).
Using single-cell genomics to study KMT2A-r ALL in samples from infants and children, the authors focus on developmental state heterogeneity, steroid resistance, lineage switching, and immune evasion. Created with Biorender.com.
Using single-cell genomics to study KMT2A-r ALL in samples from infants and children, the authors focus on developmental state heterogeneity, steroid resistance, lineage switching, and immune evasion. Created with Biorender.com.
To investigate if tumor cells resemble heterogeneous developmental states, Chen et al performed single-cell RNA-sequencing (scRNA-seq) paired with single-cell chromatin accessibility assays (scATAC-seq) on both healthy control bone marrow (n = 5) and bone marrow from patients with KMT2A-r ALL (n = 25). Using bioinformatics methods to generate a developmental trajectory for these data, the authors found that KMT2A-r ALL leukemia cells arrest at a broad range of B-cell developmental stages, which is in line with previous studies that indicate differentiation arrest at distinct stages in B-ALL subtypes.5,6 They found that blasts from young infants (<6 months) show a significantly wider range of stages within B-cell development relative to older children, including an immature population with stem-like transcription. Further, the authors found aberrant coexpression of genes from B-cell and myeloid lineages in younger infants. Together, these findings indicate that young infants with ALL exhibit more cell state plasticity compared with older children. These features have been linked to chemotherapy resistance and poor outcomes in other hematologic malignancies.5-7
Next, the authors aimed to identify the molecular regulators and functional consequences of this increased heterogeneity among younger patients. They found that steroid response genes, such as NR3C1 and KLF9 transcription factors, have lower expression, lower chromatin accessibility, and higher promoter DNA methylation in younger infants. Single-cell measurements were essential for discovering these regulators, which were not evident from bulk sequencing, because the downregulated gene signatures were specifically identified in the most immature cell populations. The authors suggested that an earlier cell of origin may cause lowered steroid responsiveness and could lead to a more resistant disease. Indeed, young infants with KMT2A-r ALL have more steroid resistance than older children, leading to poorer outcomes.8
The phenomenon of lineage switching from a lymphoid to a myeloid fate in KMT2A-r ALL has been tied to poor outcomes and predominantly occurs in the younger patient population.9,10 The authors provided evidence of lymphoid-to-myeloid lineage switch priming at the time of diagnosis in younger infants. This finding supports the model that KMT2A-r ALL originates from an early, uncommitted precursor, with a subset of patients demonstrating myeloid potential. In this study, two patients provided critical evidence that these myeloid-biased subpopulations come to the forefront under pressure by B-cell–directed immunotherapy (CART-19 or -22). Samples taken at B-ALL diagnosis (before lineage switching) showed signatures of myeloid priming, with scATAC-seq having the greatest power to detect this potential.
Finally, Chen et al gained new mechanistic insights into immune evasion, a central component of cancer development. They identified a small population of stem-like CD34+CD19− cells with KMT2A fusion transcripts. These stem-like leukemia cells are predominantly present in younger patients and express interferon (IFN) γ response signatures, alongside increased IFN γ signaling in T and natural killer (NK) cells. Most ligand-receptor interactions between stem-like cells and T/NK cells are immunosuppressive, such as killer immunoglobulin-like receptors and major histocompatibility complex class I proteins and transforming growth factor β and transforming growth factor β receptor. Functional validation confirmed that stem-like cells are resistant to NK-mediated cytotoxicity. The authors suggested that immunosuppressive signaling circuits may contribute to tumor cell persistence and explain why hematopoietic stem cell transplant is relatively ineffective in young infants with KMT2A-r ALL.
Overall, Chen et al highlighted multiple factors that may underlie poor prognosis in young patients with KMT2A-r ALL, focusing on the origin of leukemic cells, immune cell interactions, and the plasticity of blasts following current treatment strategies. These findings help explain why leukemias in young patients evade chemotherapeutic and immune-mediated approaches. Increased plasticity of leukemia cells and the expansion of immunosuppressive stem-like cells likely contributes to poor outcomes of infants with ALL. Evidence of cellular plasticity highlights the importance of targeting heterogeneous cell states, including rare populations of primitive cells. Further studies are needed to confirm and dissect mechanisms of immune evasion and/or suppression by leukemia cell subsets to design new therapeutic strategies. These efforts will be particularly rewarding, considering the potential benefit to young infants with leukemia.
Conflict-of-interest disclosure: The authors declare no competing financial interests.