TO THE EDITOR:
The syndrome of vaccine-induced immune thrombotic thrombocytopenia (VITT) is a rare thromboembolic complication of adenoviral-vectored severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines ChAdOx1 nCoV-19 (AstraZeneca) and Ad26.COV2.S (Janssen/Johnson & Johnson) mediated by antibodies directed against platelet factor 4 (PF4).1-5 The mechanisms by which the adenoviral DNA vectors break immune tolerance to PF4 and trigger B-cell clonal expansion and secretion of anti-PF4 immunoglobulin Gs (IgGs) are under intense investigation and likely involve formation of immunogenic complexes of PF4 with vaccine components in a proinflammatory setting.6 Pathogenic anti-PF4 IgGs subsequently form circulating immune complexes with PF4 tetramers, which are thought to drive thrombotic events by Fc γ receptor IIa–dependent platelet activation and to activate granulocytes to release procoagulant neutrophil extracellular traps.6-8 Serum anti-PF4 antibodies are mostly transient and appear in serum within days of vaccination, suggesting a recall immune response on memory B cells.9
Given their causal role in VITT, identification of the molecular composition of the anti-PF4 antibodies and their antigenic target(s) is crucial for better understanding of the pathogenesis and for developing better diagnostics and treatments. In a key advance, Huynh et al have mapped the antibody-binding site to a single conformational epitope on the PF4 molecule, which is located within the heparin-binding site and distinct from epitopes bound by serum from patients with heparin-induced thrombocytopenia (HIT).10 Moreover, a recent intact mass spectrometric analysis of anti-PF4 IgGs in patients with VITT and HIT revealed expression of monoclonal and oligoclonal light chains in the former as distinct from a polyclonal light-chain pattern in the latter. Although intact light-chain mass measurements were performed to inform clonality, direct amino acid sequencing of light or heavy chains was not investigated in this study.11,12
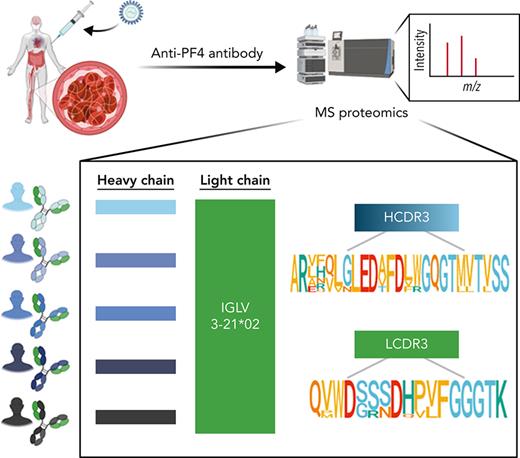
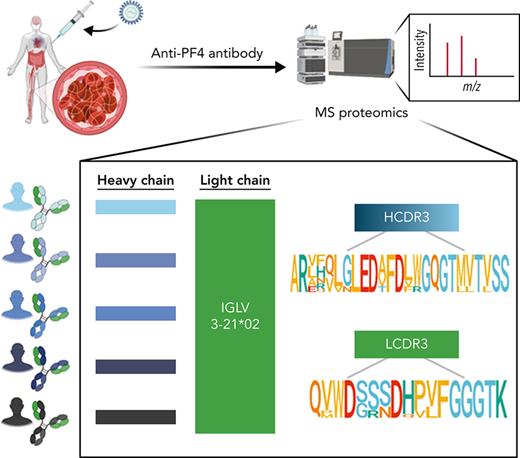
We have developed an efficient in-house proteomic workflow based on de novo mass spectrometric sequencing of immunopurified serum antibodies to identify their immunoglobulin variable (IgV) subfamily expression profiles; clonotypical light- and heavy-chain third complementarity-determining region (LCDR3 and HCDR3) amino acid sequences as barcodes for clonal tracking; and V region amino acid replacement mutational signatures as molecular markers of antigen-driven intraclonal diversification.13-16 Here, we have used a proteomics discovery approach (Figure 1A) to profile serum anti-PF4 antibodies in VITT patients and unexpectedly reveal stereotypic (also termed public) LCDR3 and HCDR3 amino acid sequences with near perfect light-chain stereotypy. This points to highly convergent pathways of anti-PF4 antibody production and potential translation of shared CDR3 peptide “barcodes” to novel molecular biomarkers for these highly pathogenic clonotypes.
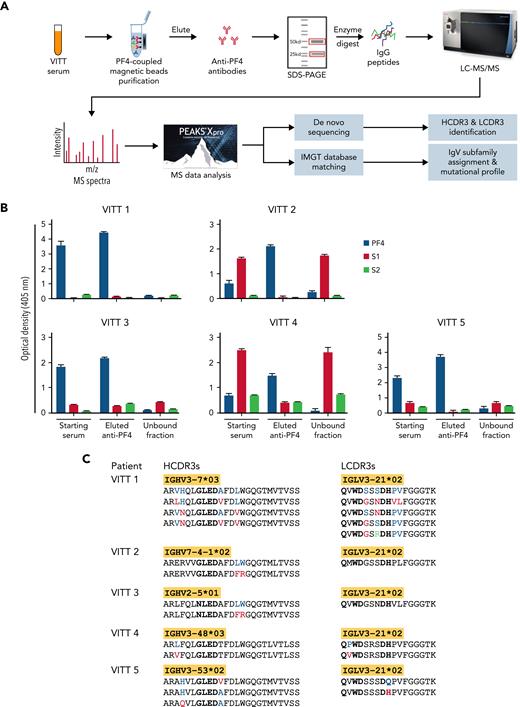
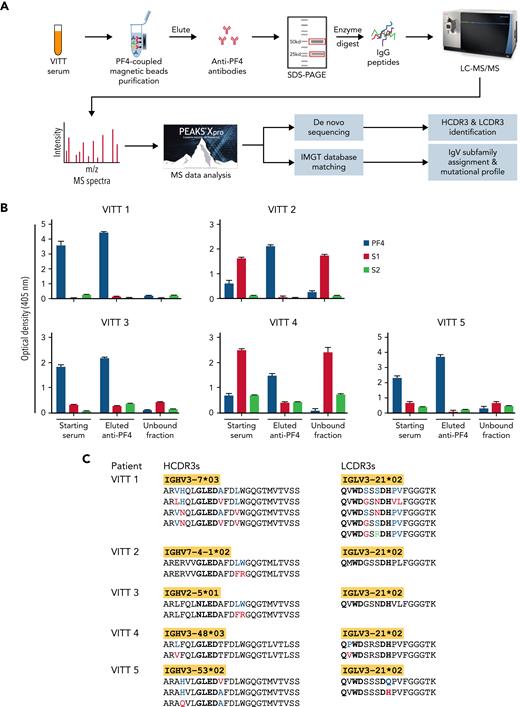
Mass spectrometry (MS)-based characterization of PF4-specific clonotypic antibodies. (A) Proteomics workflow to identify molecular signatures of anti-PF4 antibodies. PF4-specific immunoglobulins are purified from serum of VITT patients using PF4-coupled magnetic beads. Heavy (H) and light (L) chains are separated by reduced sodium dodecyl–sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and excised and digested with enzymes to generate peptides for liquid chromatography mass spectrometry/mass spectrometry (LC-MS/MS). IgV region peptide sequences are analyzed by combined de novo sequencing and IMGT database matching. (B) Specificity of purified anti-PF4 antibodies. Monospecificity of magnetic bead–purified anti-PF4 IgGs is verified by testing starting serum, bead-purified anti-PF4 antibody fraction, and unbound fractions using ELISAs coated with individual PF4, SARS-CoV-2 spike S1, and S2 proteins. Data are shown as mean ± standard deviation (n = 2). (C) Clonotypic H- and L-chain third complementarity-determining region (CDR3) signatures. IgV region subfamilies of PF4-specific antibodies as highlighted in yellow are assigned by IMGT database matching. HCDR3 and LCDR3 amino acid sequences from 5 individual VITT patients are identified by de novo sequencing. Bold amino acids in HCDR3s and LCDR3s denote shared motifs across unrelated patients, and amino acids in color denote amino acid replacement mutations found in individual patient HCDR3 and LCDR3 regions.
Mass spectrometry (MS)-based characterization of PF4-specific clonotypic antibodies. (A) Proteomics workflow to identify molecular signatures of anti-PF4 antibodies. PF4-specific immunoglobulins are purified from serum of VITT patients using PF4-coupled magnetic beads. Heavy (H) and light (L) chains are separated by reduced sodium dodecyl–sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and excised and digested with enzymes to generate peptides for liquid chromatography mass spectrometry/mass spectrometry (LC-MS/MS). IgV region peptide sequences are analyzed by combined de novo sequencing and IMGT database matching. (B) Specificity of purified anti-PF4 antibodies. Monospecificity of magnetic bead–purified anti-PF4 IgGs is verified by testing starting serum, bead-purified anti-PF4 antibody fraction, and unbound fractions using ELISAs coated with individual PF4, SARS-CoV-2 spike S1, and S2 proteins. Data are shown as mean ± standard deviation (n = 2). (C) Clonotypic H- and L-chain third complementarity-determining region (CDR3) signatures. IgV region subfamilies of PF4-specific antibodies as highlighted in yellow are assigned by IMGT database matching. HCDR3 and LCDR3 amino acid sequences from 5 individual VITT patients are identified by de novo sequencing. Bold amino acids in HCDR3s and LCDR3s denote shared motifs across unrelated patients, and amino acids in color denote amino acid replacement mutations found in individual patient HCDR3 and LCDR3 regions.
Serum specimens were obtained from 5 patients with AstraZeneca-associated VITT, and their demographic, clinical, and serological findings are summarized in Table 1. As denoted in the workflow (Figure 1A), anti-PF4 IgGs were immunopurified by PF4-coupled magnetic beads from VITT patient serum. Prior to sequencing, monospecificity of anti-PF4 IgGs was verified by ELISAs, and no cross-reactivity was found between eluted anti-PF4 IgGs and SARS-CoV-2 S1 and S2 proteins, consistent with a recent report17 (Figure 1B). Purified anti-PF4 IgGs were then separated by sodium dodecyl–sulfate polyacrylamide gel electrophoresis; heavy- and light-chain bands excised for in-gel digestion; and analysis of peptides performed in a Thermo Orbitrap Fusion Lumos Tribrid mass spectrometer with de novo sequencing and IMGT database matching. See more details for patients and methods in the supplemental methods, available on the Blood Web site.
Mass spectrometric sequencing of anti-PF4 immunoglobulins revealed a single IgG H-chain species paired with a single λ L-chain species in all 5 unrelated patients. In contrast, murine anti-human PF4 monoclonal antibodies (eg, KKO and RTO) are IgG κ.18 Remarkably, all L chains were encoded by the identical IGLV3-21∗02 gene subfamily and showed identical LCDR3 peptide lengths consistent with a high degree of L-chain stereotypy. Notably, the shared IGLV3-21∗02 allele expresses an acidic (negatively charged) DDxD motif in the CDR2 region, which we suggest may be of potential importance in antibody binding to the positively charged PF4 epitope.10 Another shared amino acid motif of interest QxWD is located in the LCDR3 region (Figure 1C). The roles of these putative binding motifs await confirmation by formal structural studies. The frequencies of different alleles of the IGLV3-21 gene vary among ethnicities, with the highest prevalence of IGLV3-21∗02 in Europeans and lowest in East Asians.19 The IGLV3-21∗02 represents ∼4% of IGLV transcripts from peripheral blood mononuclear cells in healthy donors and has been observed as a minor component of the total serum antibody proteome, such as human anti–double-stranded DNA and anti-Ro60 autoantibodies.13,15,19 To our knowledge, the dominant stereotyped expression of IGLV3-21∗02 has not been observed in any other serum antibody responses to date and can be regarded as a unique fingerprint of anti-PF4 IgGs in VITT.
HCDR3 peptides are specific clonotypic markers of serum antibodies. However, these peptides cannot be identified by conventional database matching owing to the lack of reference databases in IMGT. Accordingly, we have developed an advanced de novo sequencing workflow to identify HCDR3 clonal barcode peptides and identified striking stereotypic features characterized by identical HCDR3 lengths and homologous sequences, together with a shared binding motif G/NLED, which was located in immunoglobulin heavy chain diversity regions known to confer antigen-binding specificity20 (Figure 1C). Notwithstanding these convergent HCDR3 regions, individual patient anti-PF4 immunoglobulin proteomes were encoded by distinct immunoglobulin heavy chain variable region subfamilies, including 3-7, 7-4, 2-5, 3-48, and 3-53 (Figure 1C), emphasizing the critical role of HCDR3s in PF4 epitope binding as opposed to the divergent immunoglobulin heavy chain variable regions.
Amino acid replacement mutations were also found in individual patient HCDR3 and LCDR3 regions (Figure 1C), consistent with a model of PF4 antigen-driven intraclonal diversification as observed for systemic autoantibodies in lupus and Sjögren syndrome.13,14,21 Additional glutamic acid (E) and aspartic acid (D) replacement mutations of potential binding significance were identified in the IgV regions of H and L chains, suggesting recall immune responses on PF4-specific memory B cells (data available upon request).
The finding of a stereotyped clonotypic anti-PF4 antibody in this preliminary study of a small number of subjects represents a significant advance in elucidating the molecular pathways of pathologic antibody production in VITT and offers a rare example in human disease of a dangerous small B-cell clone that undergoes rapid clonal expansion and secretion of a harmful monoclonal antibody.22 The identification of anti-PF4 proteomic signatures using the workflow herein provides an entry point to analyze these immunological processes at a molecular level; develop novel diagnostic biomarkers for these pathogenic antibodies; develop novel therapies aimed at removing pathogenic clones (eg, anti-idiotypes and/or small peptide antibody inhibitors); and allow profiling of anti-PF4 clonal evolution invisible to current immunoassays. Moreover, the stereotyped expression of the IGLV3-21∗02 light chain paves the way for a potential genetic screening tool to identify patients carrying this gene variant who are at risk of this severe complication. Convergent anti-PF4 antibody responses are driven by selective responses to shared epitopes. Given Ad26.COV2.S and ChAdOx1 nCoV-19 vaccine-elicited anti-PF4 IgGs bind the same epitope and express monoclonal λ light chains, we anticipate they will share highly similar clonotypic signatures.11,23 Moreover, it will be of diagnostic and pathogenetic significance to compare monoclonal stereotypic anti-PF4 responses in VITT with anti-PF4/heparin antibodies in HIT. The latter binds to different epitopes on PF4 and are therefore likely to select different sets of clonotypes.10 Priorities will be to extend the study to a larger cohort and perform multi-omics characterization of linked PF4-specific serum and B-cell receptor repertoires, as reported for other human antibodies.16,24,25
Acknowledgments
The authors thank the patients for their active participation in this study.
J.J.W. is supported by a Flinders University DVCR Fellowship and a Flinders Health & Medical Research Institute COVID-19 Research Grant.
Authorship
Contribution: J.J.W. and T.P.G. developed the study concept, designed the study, compiled the data, performed the analyses, and wrote the paper; B.A. performed the experiments, participated in designing the research, analyzing the data, and writing of method; T.C. participated in acquisition, analysis, and interpretation of data; A.T. participated in study design and interpretation of data; A.C. performed acquisition of data and participated in writing of method; O.Y. recruited and enrolled the patients and performed patient sample collection and diagnostic assays; S.H. participated in developing the study concept and recruiting patients; C.W.T. recruited and enrolled the patients, provided patient information, and participated in designing the research, interpreting the data, and writing the paper; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jing Jing Wang, Department of Immunology, College of Medicine and Public Health, Flinders University, Bedford Park, SA 5042, Australia; e-mail: jingjing.wang@flinders.edu.au.
References
Author notes
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.