In this issue of Blood, Cunningham et al1 use an extensive array of metabolic depletion and supplementation experiments across several complementary models to identify a selective dependence of acute myeloid leukemia (AML) cells on the essential amino acid methionine (Met).
Metabolic reprogramming is an established cancer hallmark, and particularly in AML, it has been shown to play a role in disease establishment, progression, and therapy resistance.2 Therefore, identifying AML-specific metabolic vulnerabilities that can be targeted therapeutically has become an area of active research. Among many metabolic pathways that are reprogrammed in AML, amino acid metabolism has attracted particular attention. This is because differential amino acid dependencies have been identified between normal hematopoietic stem/progenitor cells (HSPCs) and their leukemic counterpart, including leukemia stem cells (LSCs) and therapy-resistant AML cells. Indeed, normal HSPCs specifically rely on the branched chain amino acid valine3 for their proliferation and maintenance, whereas LSCs and therapy-resistant AML cells have been shown to rely on different amino acids, such as cysteine4 and glutamine5 among others, to fuel their tricarboxylic acid cycle and oxidative phosphorylation metabolism and to withstand therapeutic pressure.
Cunningham et al add another piece to the jigsaw puzzle of the functional role of specific amino acids in AML by showing that dietary Met restriction specifically depletes AML cells while sparing normal HSPCs. The authors first screened the effects of individually depleting each proteinogenic amino acid on the growth of AML cell lines and primary AML samples in vitro. Not only could they recapitulate previous findings on cysteine dependency in AML,4 but they also identified Met depletion across all their models as a strong and selective dependence of AML cells. They then showed that Met restriction in vivo reduced leukemic burden and prolonged survival in both cell line and patient-derived xenografts. Interestingly, normal HSPCs were only marginally affected by Met depletion, with no effects on their viability in vitro or frequencies in vivo.
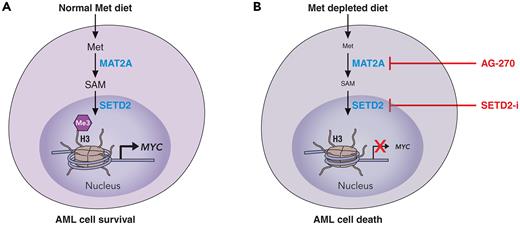
To elucidate the mechanistic underpinning of their striking phenotypic observations, Cunningham et al embarked in a meticulous series of experiments supplementing individual metabolites from the Met cycle and transsulfuration pathway in Met-free culture conditions. Through this work, they were able to show that S-adenosylmethionine (SAM) was the only metabolite able to rescue the effects of Met depletion. SAM is produced from Met through the activity of the MAT2A enzyme, and indeed, the authors showed that the effects of Met depletion were phenocopied when they used a specific MAT2A inhibitor. Because SAM is a universal methyl donor used by multiple histone methyltransferases, the authors then directed their attention to changes in histone methylation in cells grown in Met-free media. They showed that Met depletion mostly reduced levels of trimethylated histone H3 at lysine 36 (H3K36me3) in a SAM-dependent manner and preferentially in AML cells. H3K36me3 is a mark present on actively transcribed genes, and Met depletion led to selective reduction of H3K36me3 abundance at genes involved in protein translation, cell cycle, and antiapoptosis such as MYC and BCL2L2. As a result, Met depletion led to a defective protein translation in AML cells mostly affecting proteins with key functions in survival and proliferation, thus explaining the mechanism underlying the antileukemic effects of Met depletion. Indeed the authors also could mimic the effects of Met starvation by chemically inhibiting SETD2, the specific H3K36 trimethyl transferase, further supporting the specific role of reducing H3K36me3 levels in driving AML cell death after Met depletion (see figure).
Effects of Met dietary depletion on AML cells. (A) In normal Met dietary replete conditions, AML cells are able to metabolize Met through various steps to SAM to support histone 3 lysine 36 trimethylation and transcription of key genes involved in leukemia cell survival and proliferation. (B) On Met depletion, or through pharmacologic targeting of the enzymatic steps that lead to the formation of SAM from Met, AML cells are unable to support their survival transcriptional programs, which in turn leads to cell death. Professional illustration by Patrick Lane, ScEYEnce Studios.
Effects of Met dietary depletion on AML cells. (A) In normal Met dietary replete conditions, AML cells are able to metabolize Met through various steps to SAM to support histone 3 lysine 36 trimethylation and transcription of key genes involved in leukemia cell survival and proliferation. (B) On Met depletion, or through pharmacologic targeting of the enzymatic steps that lead to the formation of SAM from Met, AML cells are unable to support their survival transcriptional programs, which in turn leads to cell death. Professional illustration by Patrick Lane, ScEYEnce Studios.
Although considered an essential amino acid, Met can be recycled from homocysteine. Interestingly, findings from the work of Cunningham et al suggest that in many AML cells, this recycling may be defective compared with healthy HSPCs because of the reduced expression of the enzymes involved in this pathway. This in turn may explain the enhanced sensitivity of AML cells to Met depletion. Whether the activity of this Met recycling pathway can also be used as a biomarker of response to Met depletion in AML or could be targeted to enhance the effects of Met deprivation requires further studies.
Overall, this work provides a strong rationale to target Met metabolism in AML. The mechanistic insight clearly shows that rewired metabolism feeds into aberrant transcription, another hallmark of AML,6 and adds to the growing literature showing how metabolism and epigenetic dysregulation are interlinked closely in cancer and represent 2 sides of the same coin that can be actioned therapeutically to eradicate cancer cells.7
Regarding how to best leverage the Met dependence of AML cells, it is worth noting that Met restricted diets already have been trialed in humans with good tolerability while achieving significant reduction in plasma levels of Met and related metabolites.8 Such an approach may be a more tolerable, less expensive, and possibly more effective way to achieve therapeutic benefit. Despite some general concerns about compliance, evidence is growing that dietary interventions can be implemented safely and effectively in cancer patients.9 Alternatively, targeting the key nodes in Met metabolism in AML cells, such as MAT2A and SETD2, via small-molecule inhibitors may provide a more conventional therapeutic approach.
Although specific mutations can dictate selective metabolic vulnerabilities, as is the case in isocitrate dehydrogenase mutant AML,10 often metabolic dependencies transcend specific AML genetic backgrounds. Therefore, a significant challenge in the clinical implementation of metabolic therapies is the need to understand better how metabolic dependencies become truly lethal metabolic bottlenecks. This requires the identification of validated biomarkers predictive of therapy response. Alternatively, metabolic perturbations may unmask vulnerabilities that can be leveraged through ancillary therapeutic interventions in a true synthetically lethal fashion, and these could be exploited to improve treatment outcomes. Only through such efforts can the AML community hope to fulfil the promise of delivering more effective, less toxic metabolic-targeted therapies in AML in a true precision medicine approach. The work from Cunningham et al provides an important step toward achieving this goal.
Conflict-of-interest disclosure: The author declares no competing financial interests.