Abstract
Max-gene associated (MGA) is a dual-specificity transcription factor that is recurrently mutated in 2-5% of all cancers and commonly seen in lung adenocarcinoma, endometrial carcinoma, and colorectal cancer. Our group has previously described heterozygous loss-of-function mutations in 9% of RUNX1::RUNX1T1 pediatric AMLs. Similar loss-of-function mutations are seen in other hematopoietic malignancies, including chronic lymphocytic leukemia (CLL), natural killer/T-cell lymphoma, B-cell acute lymphoblastic leukemia (B-ALL), and T-cell acute lymphoblastic leukemia (T-ALL). MGA is a component of the non-canonical polycomb repressive complex (ncPRC1.6) containing an amino-terminal T-box domain and a carboxy-terminal Myc-like basic helix-loop-helix (bHLH) domain which are used to regulate MAX-network and T-box family targets. In hematological malignancies such as RUNX1::RUNX1T1 AMLs, MGA mutations disrupt or remove the Myc-like bHLH domain. The role of MGA in normal hematopoietic regulation and leukemic development are understudied. This is likely due to the lack of in vivo MGA deficiency models which are embryonic lethal. We hypothesize that the loss of MGA leads to disruption of the ncPRC1.6 complex, causing a loss in global transcriptional repression of many pathways, including Myc and E2F target genes, leading to enhanced hematopoietic cell growth and self-renewal, as well as shortening the latency of AML driven by RUNX1::RUNXT1. To test this hypothesis, we designed a multidisciplinary approach to characterize the hematopoietic function of MGA.
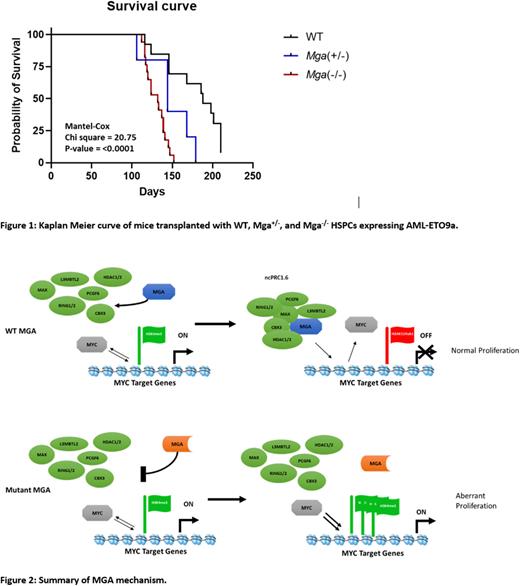
IP-Westerns of overexpressed flag-tagged MGA and its subsequent variants in K562 cells revealed that MGA truncations disrupt interactions with components of the ncPRC1.6 complex. Because these truncations lead to the loss of MGA's molecular function, we next generated CRISPR/Cas9 to knockout MGA in Molm13 cells. Using RNA-Seq and CUT&RUN to assess the genomic localization of MGA, we determined that MGA binds to an average of 85,119 peaks and 17.7% of those sites are at transcription start sites, including cell cycle genes such as CCND2. Deletion of MGA led to the removal of repressive H3K27me3 marks and an increase in MYC binding as well as active histone marks such as H3K4me3 and H3K27Ac at the transcription start site of CCND2. This led to an increase in CCND2 expression (logFC 1.56, FDR 1.56E-12) which is supported by in vitro EdU assays showing a 10.43% increase in cells cycling into S-phase. To further define the hematopoietic function of MGA, we designed a conditional knockout Mga mouse model with LoxP sites flanking exon 3, leading to deletion of Mga in hematopoietic stem and progenitor cells (HSPCs) when crossed with Vav1-cre. Transcriptome profiling analysis of HSPCs isolated from heterozygous (Mga+/) and homozygous (Mga-/-) mice reveals that Mga-/- mice, but not Mga+/-, leads to up-regulation of cell cycle (FDR 4.2E-06), Myc (FDR 6.3E-10), and E2f (FDR 2.9E-17) signaling pathways. Ex vivo serial CFU replating and cell proliferation assays support this, showing that Mga-/- leads to increased stem cell self-renewal and cell growth. However, bone marrow and spleen analysis of both Mga+/- and Mga-/- mice reveal no changes in immunophenotype when compared to WT controls. Expression of RUNX1::RUNX1T1 isoform 9a (AML-ETO9a) in Mga+/- or Mga-/- HSPCs reveals a significant increase (69% and 74% respectively, p<0.01) in leukemic stem cell self-renewal, when compared to WT controls expressing AML-ETO9a at week 4. When transplanted in mice, Mga+/- and Mga-/- HSPCs expressing AML-ETO9a led to a rapid expansion of blasts (GFP+, lin-, cKit+) found in the peripheral blood as well as a shortened latency compared to wild-type controls expressing AML-ETO9a (Figure 1).
These studies demonstrated MGA as a regulator of hematopoietic cell proliferation (Figure 2), and how MGA deficiency cooperates with oncogenes to create more aggressive leukemia. Because MGA mutations occur in many tumor types, these data also provide functional insights into ncPRC1.6 activity in leukemia and other tumor types with recurrent mutations in MGA or other components of this complex.
Disclosures
No relevant conflicts of interest to declare.
Author notes
*Asterisk with author names denotes non-ASH members.