Abstract
Introduction
Genetic testing is frequently required for aiding and confirming the diagnosis, selecting treatment and predicting prognosis in patients with hematological disorders (1). However, large-scale genetic studies based on whole exome or genetic panel sequencing have not been a primary focus of developing countries due to lack of awareness, a dearth of genetic testing laboratories in clinical facilities, and the limited financial resources of affected families. As a result, the underlying genetic patterns in these patients often go undetected. Nevertheless, these countries constitute a rich source of clinical data with atypical phenotypes and novel mutations due to large family sizes, inter-family marriages, and potential founder effects (2). These genotype and phenotype data are worth being presented in the international data repositories for better identification of pathogenic variants in this population (3). Armed Forces Bone Marrow Transplant Centre (AFBMTC) is the country's largest hospital providing clinical hematology and stem cell transplantation services to a catchment area of over 560,000 Km2. We sought to identify underlying genetic causes of various difficult-to-diagnose rare hematological disorders with heterogenous/overlapping symptoms and challenging clinical presentation using next-generation sequencing (NGS).
Materials and Methods
Peripheral blood samples from 33 individuals with diverse hematological diseases were sent to Invitae Corporation (USA) or Centogene GmbH (Germany) for panel sequencing or whole exome sequencing, respectively. These samples were from patients who were difficult-to-diagnose, required risk stratification or genetic testing for selecting appropriate treatment.
Results
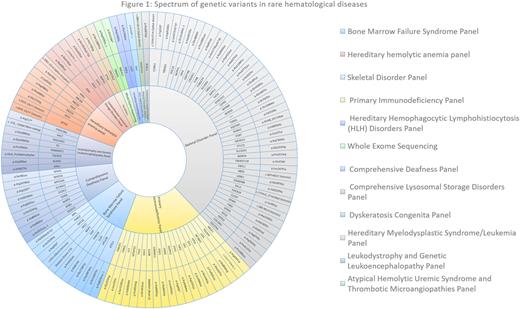
Whole exome sequencing or panel sequencing was performed in a cohort of 33 patients with various hematological disorders (figure 1). A total 115 variants were identified in these patients including 15 pathogenic variants (13%), 7 likely pathogenic variants (6%), 82 variants of uncertain significance-VUS (71%), 9 benign variants (7.8%) and 1 increased risk allele variant (0.86%). A total of 42 variants were found in musculoskeletal disorders panel, 20 in primary immunodeficiency panel, 11 each in bone marrow failure syndrome panel and dyskeratosis congenita panel, 9 in comprehensive deafness panel and 9 in hereditary hemolytic anemia panel among others. The most frequent diagnosis in our cohort was inherited bone marrow failure syndromes (27.3%), followed by hemolytic anemias (15.2%), immunodeficiency disorders (12.2%), inherited platelet function disorders (12.2%), myelofibrosis disorders (12.2%), skeletal disorders (12.2%) and miscellaneous disorders (9.1%). Patients clinical and demographic characteristics are summarized in table 1.
Conclusion
Use of high-throughput NGS in our clinical hematology setting has proven to be useful in identifying genetic variants for accurate diagnosis, selecting appropriate therapies, and predicting prognosis. We intend to develop a genetic facility that will provide genetic testing and identify ways to make it more accessible for people of all socioeconomic backgrounds. A second objective is establishing the prevalence of most frequent genetic disorders in the country. The high number of variants of uncertain significance (VUS) is most likely driven by a lack of sufficient population-based genetic data and proof of functional impact, though the patients identified having VUS were phenotypically disease presenting. Thus, supporting the hypothesis that these VUS may eventually be classified as pathogenic after appropriate testing on larger scales and animal models. To achieve a dependable and precise categorization of VUS, we plan to include functional genomics in our research focus to support the clinical database of this region.
1. Haferlach TJh. The time has come for next-generation sequencing in routine diagnostic workup in hematology. 2021;106(3):659.
2. MALTESE P, Poplavskaia E, Malyutkina I, Sirocco F, Bonizzato A, Capodicasa N, et al. Genetic tests for low-and middle-income countries: a literature review. 2017;16(1).
3. Gutierrez-Rodrigues F, Calado RTJH, Transfusion, Therapy C. The interpretation of rare or novel variants: Damaging vs. disease-causing. 2018;40:3-4.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.