Abstract
Background: Janus kinase 2 (JAK2) is a non-receptor tyrosine kinase that activates transcriptional programs of cell survival and proliferation, through signal transducers and activators of transcription (STAT) family of transcription factors, in response to stimulation by growth factors and cytokines. Upregulation of JAK/STAT signaling is common in myeloproliferative neoplasms (MPNs), which are also clinically associated with accelerated catabolism (weight loss, exhaustion). JAK2 inhibitors can improve these symptoms and improve quality of life for patients with MPNs, but they are unable to eradicate disease-initiating malignant hematopoietic stem cells, indicating that our understanding of the molecular pathophysiology of MPNs is incomplete and that new therapeutically targetable mechanisms need to be uncovered to improve therapy.
Objective: To better understand signaling patterns implicating JAK2 V617F in malignant transformation, we interrogated the JAK2 WT and JAK2 V617F cellular interactomes using proximity dependent labeling with biotin (PDL) and mass spectrometry.
Methods: Retroviral (MSCV-IRES-GFP) vectors expressing JAK2 WT or JAK2 V617F fused with biotin ligase (TurboID, PMID: 30125270) were established and used to transduce murine myeloid FDC-P1 cells. FDC-P1 cell lines stably expressing TurboID alone, TurboID-JAK2 WT, or TurboID-JAK2 V617F were incubated with 50 μM biotin for 3 hours. Cells were lysed and biotinylated proteins were pulled down using streptavidin conjugated beads. On-bead trypsin digestion followed by mass spectrometry were performed to determine a pool of biotinylated proteins representing a network of proximal interactors of TurboID, TurboID-JAK2 WT, or TurboID-JAK2 V617F. Significantly enriched proximal interacting proteins identified in TurboID-JAK2 V617F or TurboID-JAK2 WT pulldowns were designated as having 1.5-fold more enrichment (FDR ≤ 0.05) over TurboID, and showing 1.5-fold difference in enrichment (FDR ≤ 0.05) between TurboID-JAK2 V617F and TurboID-JAK2 WT.
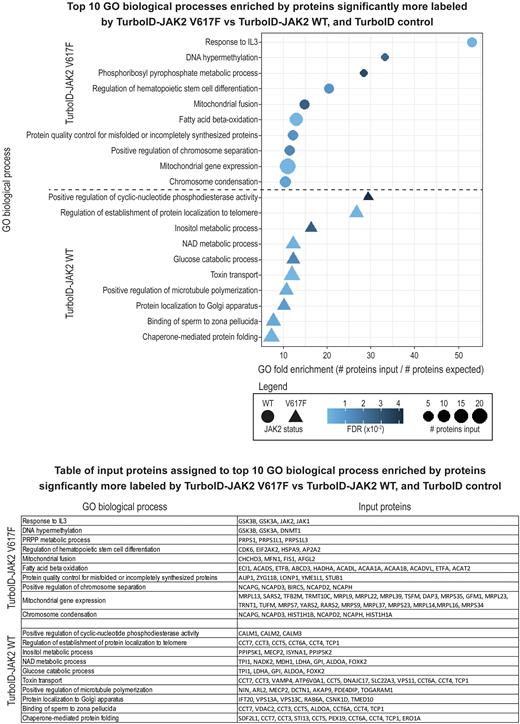
Results: 441 proteins were identified to be significantly enriched in TurboID-JAK2 WT over TurboID control and significantly enriched over TurboID-JAK2 V617F. 330 proteins were significantly enriched in TurboID-JAK2 V617F over TurboID control and significantly enriched over TurboID-JAK2 WT. Bioinformatic analyses of proteins more enriched by TurboID-JAK2 WT or TurboID-JAK2 V617F both identified hematopoietic system as a significant tissue, and nucleoside triphosphatase activity as a significant gene ontology (GO) molecular function. Top 10 GO biological process terms enriched exclusively in TurboID-JAK2 WT included "Positive regulation of establishment of protein localization to telomere", and "Glucose catabolic process" (Panel 1). GO biological process terms exclusively enriched in TurboID-JAK2 V617F included "Cellular response to interleukin-3", "Fatty acid beta-oxidation", and "Mitochondrial gene expression" (Panel 1). See table in Panel 1 for list of proteins assigned to significantly enriched GO biological process terms.
Conclusions: Our data indicate that JAK2 V617F is associated with changes in cellular energy metabolism, characterized by increased association with proteins involved in fatty acid beta-oxidation, and decreased association with proteins involved in glucose metabolism. These data are consistent with the report of Skoda group showing that expression of JAK2 V617F resulted in systemic metabolic changes including hypoglycemia, adipose tissue atrophy, and early mortality (PMID: 31511238). Our study identifies new potential JAK2 V617F interactors involved in regulation of fatty-acid oxidation and mitochondrion linked processes. Further studies are warranted to determine druggability of the newly identified proximal interactors to revert JAK2 V617F-dependent pathologies.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.