Abstract
Objective: To investigate the prevalence of gene variation and related survival in childhood acute myeloid leukemia (AML) in South China.
Methods: A retrospective analysis was performed on 546 children with AML (non-APL) admitted to the nine hospitals in South China from January 2015 to December 2020. The gene mutation spectrum was detected by first or next-generation gene sequencing. All of patients received C-HUANAN AML 2015 protocol to treatment. The induction regimen were non-randomized to DAE group and FLAG-IDA group. The induction regimen of DAE group including coures 1 (DAE 3+5+10), course 2 (DAE 3+5+8) . The induction regimen of FLAG-IDA including two courses of FLAG-IDA. Consolidation treatment was same and consisted of 2 courses: 1.HAE or MACE; 2.MidAC. Follow-up observation was started after 4 courses of treatment. The gene variation and related prognosis were analyzed.
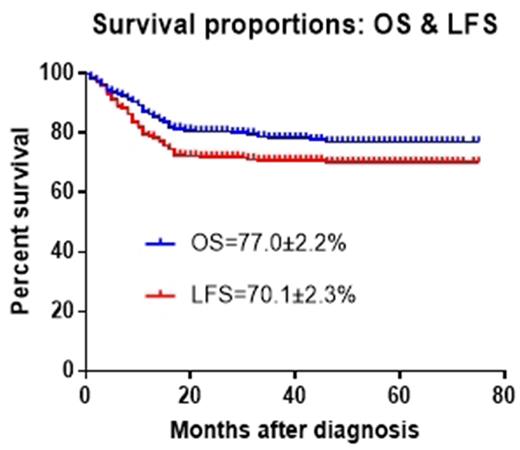
Results: A total of 546 patients with AML were included in this study, with a male to female ratio of 1.47:1, a median survival time was 71 (2-201) months , and a follow-up until April 2021, with a median follow-up time of 21 (1-75) months. Four hundred and forty eight patients survived, 98 died, and 42 died of recurrence of leukemia. The OS=77.0±2.2% and EFS=70.1±2.3% in this study.A total of 343 fusion genes and 468 mutated genes were detected, and the incidence rates of fusion genes and mutated genes were 58.1% and 54.2%. For whom with fusion gene, overall survival (OS) and event-free survival (EFS) as follows: AML1-ETO 145 cases (26.6%) ,OS: 87.0±3.4%, EFS: 85.0±3.6%. KMT2A rearranged 100 cases (18.3%) ,OS: 72.4±6.5%, EFS: 73.1 ±7.8%. while MLL-AF9 44 cases (8.1%), OS: 79.3±13.0%, EFS: 73.3± 12.7 %. CBFβ/MYH11 37case(6.8%), OS: 90.4 ±6.7%, EFS: 90.4±6.7%. For whom with mutated gene, OS and EFS as follows: C-KIT 61 cases (11.2%) OS: 89.7±5.0%, EFS: 89.7±5.0%. ASXL1 58 cases (10.6 %), OS: 80.1±7.9%, EFS: 74.7±7.4%. Fiftiy five cases of FLT3-ITDhigh (10.1%), OS: 54.4±10.3%, EFS: 54.8±10.2%.Thirty eight cases of EVI1(7.0%), OS: 56.5±12.4%, EFS: 56.5±12.4%.Seventeen cases of biallelic mutated CEBPA (3.1%), OS: 83.3±10.8%, EFS: 81.8 ±11.6%. Twelve cases of NPM1(2.2%), OS : 90.9±8.7%, EFS: 90.9± 8.7 %. The survival analysis of 363 patients who received chemotherapy only showed that the prognosis of positive AML1-ETO gene was significantly better than that of the negative of this gene (OS: 87.0 ± 3.4% vs 72.4±3.6% (χ2 =6.340 P=0.012), EFS: 85.0±3.6% vs 68.0±3. 4% (χ2=9.434 P=0.002));The prognosis of positive CBFβ-MYH11 gene was significantly better than that of negative gene (EFS: 90.4±6.7% vs 72.3±2.8 % (χ2 =3.985 P=0.046)). The survival of EVI1 gene mutation was significantly worse than that of the negative gene(OS: 56.5 ±12.4 % vs 78.6±2.6% (χ2 = 5.97 5 P=0.015)). The survival of patient with c-KIT gene mutation was significantly better than who without this gene. (EFS: 89.7±5.0% vs 71.6±2.8% (χ2 =4.292 P=0.038)). Total 155 patients whom in high risk group and intermediated risk group underwent HSCT. OS and LFS of Patients in high risk group who underwent HSCT could reached to 75.8±6.7% and 74.9±7.0% respectively while OS and LFS of patients in high risk group who only underwent chemotherapy was 54.9±7.3%(P=0.003)and 51.4±7.3% (P=0.003)respectively. Patients combined with FLT3-ITD mutation underwent HSCT got markly better OS and EFS as 81.0±10.0% and 78.8±11.0% versus 54.8±10.2% (P=0.0029)and 54.4±10.3% (P=0.028)who only received chemotherapy.
Conclusion: The multicenter study showed that the most common fusion gene in these 546 children with AML was AML1-ETO, KMT2A-rearrangement and the CBFβ-MYH11 respectively. The most common mutant gene in was WT1.,C-Kit, ASXL1 and FLT3-ITD respectively. Althought only underwent 4 courses of chemotherapy, the OS and LFS could be reached to 77.4±2.6% and 73.7±2.6%. AML1-ETO and CBFβ-MYH11 genes were favorable prognostic factors,even combined with C-Kit mutation. Patients with FLT3 - ITD and EVI1 genes had poor survival. Hematopoietic stem cell transplantation improved the survival of patients in high-risk group or with FLT3-ITD mutation.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.