Abstract
Directed differentiation of pluripotent stem cells (PSCs) into functional hematopoietic stem cells (HSCs) remains elusive. A better understanding of HSC development, both intrinsically and extrinsically, will provide insights to guide PSC-HSC differentiation. During mouse embryogenesis, a subpopulation of endothelial cells in the aorta-gonad-mesonephros (AGM) region are specified at embryonic day (E) 9.5 to hemogenic endothelial cells (HECs). HECs then undergo an endothelial to hematopoietic transition (EHT) between E9.5 and E10.5. Among these hematopoietic cells, only a few are nascent HSCs (pre-HSCs) that migrate to the fetal liver for maturation and expansion. Recent advances in single cell RNA sequencing (scRNA-seq) substantially improved our understanding of intrinsic cellular programs of human and mouse HSC development; however, much less is known about the hematopoietic niche cells and niche-emanated signals in the AGM region and fetal liver.
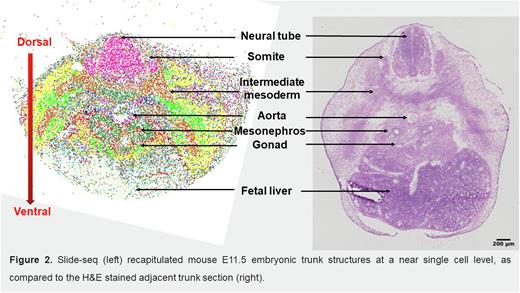
To characterize the niche components and related signaling of HSC development, we conducted a spatial transcriptomics analysis using Slide-seq (with 10 µm resolution) on mouse E10.5, E11.5 and E12.5 trunk tissues covering both the AGM region and fetal liver. We also performed scRNA-seq on seven samples collected from trunk tissues, including E10.5, E11.5 and E12.5 AGM regions; E10.5, E11.5 and E12.5 trunks; and E12.5 fetal liver. We developed an algorithm to integrate the Slide-seq and scRNA-seq data that takes advantage of their spatial and in-depth transcriptional sequencing capacity, respectively. Using E11.5 Slide-seq data as an example, we successfully identified most, if not all, of the tissue structures at a near single cell level in trunk tissue (see Figure). Furthermore, we could determine the expression of thousands of genes on individual beads, providing an invaluable tool for investigating spatial-characterized niche cells and signals. Compared to the dorsal aorta, which was dominated by one cell cluster expressing Nfgr, a complex and multilayered cellular architecture was revealed in the ventral part of aorta where EHT occurs, including pre-HSCs, endothelial cells, neural crest derived cells, mesonephric cells, and macrophages. We found a rare population of mesenchymal stromal cells (MSCs) that co-express Cdh2 (encodes N-cadherin or N-cad) and Pdgfra. Of note, these N-cad+ MSCs exclusively localize to the ventral part of the aorta as a single cell layer and are sandwiched between an inner layer of aortic endothelial cells and an outer layer of mesonephric cells. Secondly, we identified multiple genes enriched in the ventral aorta compared to the dorsal aorta, including Dlk1, Wnt4, Igf1, and Ptn. Gene Ontology analysis and KEGG signaling pathway analysis were also performed to investigate the biological processes and signaling pathways enriched in individual cell clusters. Potential signaling modules representing ligand-receptor interactions between niche cells and HECs/pre-HSCs were analyzed by CellphoneDB; the identified spatial location of niche cells relative to pre-HSCs allows us to exclude predicted interactions in which niche cells were located too far from pre-HSCs to be relevant. As a result, we generated a list of known and potentially novel signals critical for EHT, including DLK1, JAG1 and TGFβ emanating from N-cad+ MSCs, endothelial cell-derived JAG2 and DLL1/4, mesonephric cell-secreted WNT2b, GREM1 (a BMP antagonist) and R-spondin1, gonad cell-derived SPP1, and macrophage-produced TNFα. A thorough analysis on both AGM and fetal liver at different developmental stages is ongoing.
In summary, we dissected dynamic and near single cell-resolved spatial characteristics of the niche components and signals for HSC development in the AGM region and fetal liver. These findings will deepen our knowledge of embryonic hematopoiesis and guide the in vitro induction of human PSCs to functional HSCs.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.