Key Points
The mechanosensory ion channel Piezo1 is the carrier molecule for Er red cell antigens, establishing a new blood group system.
Antibodies directed against 2 novel high-incidence Er antigens are associated with severe hemolytic disease of the fetus and newborn.
Abstract
Despite the identification of the high-incidence red cell antigen Era nearly 40 years ago, the molecular background of this antigen, together with the other 2 members of the Er blood group collection, has yet to be elucidated. Whole exome and Sanger sequencing of individuals with serologically defined Er alloantibodies identified several missense mutations within the PIEZO1 gene, encoding amino acid substitutions within the extracellular domain of the Piezo1 mechanosensor ion channel. Confirmation of Piezo1 as the carrier molecule for the Er blood group antigens was demonstrated using immunoprecipitation, CRISPR/Cas9-mediated gene knockout, and expression studies in an erythroblast cell line. We report the molecular bases of 5 Er blood group antigens: the recognized Era, Erb, and Er3 antigens and 2 novel high-incidence Er antigens, described here as Er4 and Er5, establishing a new blood group system. Anti-Er4 and anti-Er5 are implicated in severe hemolytic disease of the fetus and newborn. Demonstration of Piezo1, present at just a few hundred copies on the surface of the red blood cell, as the site of a new blood group system highlights the potential antigenicity of even low-abundance membrane proteins and contributes to our understanding of the in vivo characteristics of this important and widely studied protein in transfusion biology and beyond.
Introduction
To date, the International Society of Blood Transfusion (ISBT) recognizes 378 blood group antigens, of which 345 fall into one of 43 recognized blood group systems.1 Only a small number of antigens, of both high and low incidence, remain genetically uncharacterized. Antigens that are related serologically, biochemically, or genetically but do not fall into any recognized blood group system are grouped together into collections (the 200 series).1 The Er blood group collection represents one such group of antigens, for which the carrier molecule has yet to be identified. The high-incidence Era antigen (208001) was first identified in 1982,2 with its antithetical low-incidence antigen, Erb (208002), identified 6 years later.3 The third antigen in the Er collection, Er3 (208003), is defined by an antibody produced by an Er(a–b–) individual and was proposed to represent a null phenotype.4 Although anti-Era and anti-Erb have not been implicated in either hemolytic transfusion reactions (HTR) or hemolytic disease of the fetus and newborn (HDFN), anti-Er3 production has been associated with possible mild HTR and may be clinically significant.4 However, clinical data are sparse due to the rarity of these antibodies, so conclusions about clinical significance are difficult to draw.
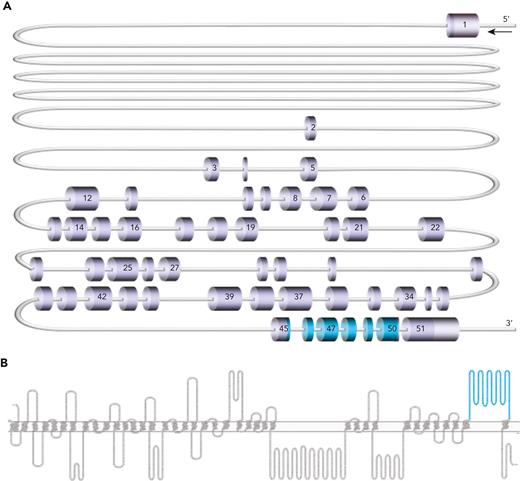
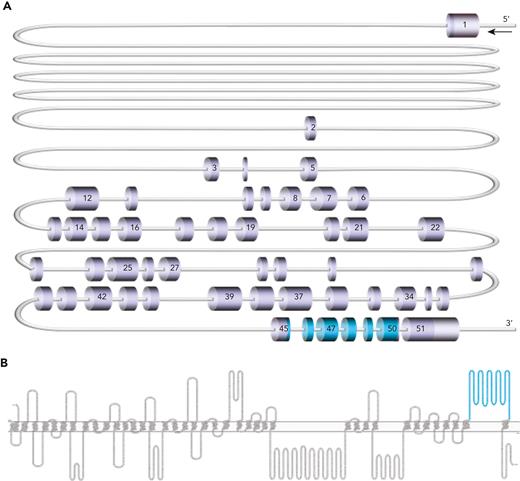
Piezo1 is a large mechanosensitive ion channel that acts as a nonselective cation channel in a variety of tissues.5,6 It is encoded by the PIEZO1 gene, comprising 51 exons and located on chromosome 16q24.3.5,7 The protein consists of 36 transmembrane domains and 1 large extracellular domain (amino acids 2198-2431), encoded by exons 45 to 50 (Figure 1), present in the membrane as a homotrimer.8 Piezo1 is involved in a host of crucial processes in the lungs, bladder, colon, and kidney, as well as in sensing of blood flow in the vasculature system.9-12 It is expressed in erythrocyte membranes,13 where it plays a vital role in regulation of red blood cell volume during circulation.14 Rare loss-of-function mutations in PIEZO1 have been shown to result in generalized lymphatic dysplasia of Fotiou (GLDF; MIM 616843), inherited in an autosomal recessive manner.15 More common, autosomal-dominant, gain-of-function mutations are associated with dehydrated hereditary stomatocytosis (DHS; MIM 194380). DHS presents with abnormal red cell morphology13,16,17 and metabolism,18 in addition to defects in red cell volume homeostasis,13,16,17 erythroid differentiation,19 reticulocyte maturation,20 and macrophage phagocytic activity.21 Both GLDF and DHS are associated with nonimmune fetal hydrops and/or perinatal edema15,22; however, the pathogenic mechanism is poorly understood.23
PIEZO1 gene and Piezo1 protein cartoons. (A) Scale representation of PIEZO1 gene (NM_001142864.4), highlighting exons 45 to 50 (shown in blue), which encode the large extracellular loop (amino acids 2198-2431) of the Piezo1 protein. (B) Piezo1 protein (Uniprot Q92508) cartoon generated in Protter24 with large extracellular loop (amino acids 2198-2431) shown in blue.
PIEZO1 gene and Piezo1 protein cartoons. (A) Scale representation of PIEZO1 gene (NM_001142864.4), highlighting exons 45 to 50 (shown in blue), which encode the large extracellular loop (amino acids 2198-2431) of the Piezo1 protein. (B) Piezo1 protein (Uniprot Q92508) cartoon generated in Protter24 with large extracellular loop (amino acids 2198-2431) shown in blue.
Although it plays a crucial role in mammalian viability (mice deficient in Piezo1 die in utero)12 and red blood cell integrity (lineage specific Piezo1 knockout mouse red blood cells are overhydrated and fragile),25 the Piezo1 protein is present in only a few hundred copies per red blood cell26 providing challenges for both detection and functional manipulation.
In this study, we identify the red blood cell ion channel Piezo1 as the elusive carrier molecule for the Er blood group antigens. Using exome sequencing of individuals with serologically defined Er-related alloantibodies, we demonstrate missense and nonsense mutations in PIEZO1, segregating with Er phenotype. Through CRISPR/Cas9-mediated gene knockout in an erythroid cell line, expression studies, proteomics, electrophysiology, and extensive serological characterization, we define the molecular bases of the currently recognized antigens of the Er collection (Era, Erb, and Er3), establishing Er as a new blood group system. We also provide evidence for 2 further high-incidence antigens of the Er system (described here as Er4 and Er5) and present data regarding clinical significance of anti-Er4 and anti-Er5.
Methods
Samples
Blood samples were procured, and the study was conducted according to NHS Blood and Transplant (NHSBT) Research and Development governance requirements and ethical standards and in accordance with the Declaration of Helsinki. Ethics approval was granted by National Health Service Health Research Authority, Bristol Research Ethics Committee reference 12/SW/0199.
Samples from 6 unrelated individuals serologically typed as Er(a–b+), 3 individuals typed as Er(a+b+), and 1 individual typed as Er(a–b–) were obtained from IBGRL cryopreserved reference collections. A further 3 individuals were included due to variant expression of Er antigens and/or identification of apparently Er-related antibodies in their serum, together with a total of 15 family members of these individuals (Table 1; supplemental Table 1, available on the Blood website).
Ethylenediaminetetraacetic acid (EDTA) peripheral blood samples from voluntary NHSBT blood donors, who consented to the use of their blood for research purposes, were used as controls.
Serologic testing
Standard agglutination techniques were used for investigation of reactivity of all Er-related antibodies and red blood cell (RBC) phenotyping using anti-Era (from P1, P2, P3, and P6), anti-Erb (IBGRL reference collection), and anti-Er3 (from P10). For the indirect antiglobulin test, a low-ionic strength saline tube method was used, where the secondary antibody was polyspecific antihuman globulin (Millipore). Anti-Er eluates were prepared using Gamma ELU-KIT II rapid acid elution kit (Immucor). Agglutination was scored on a scale of 0 (negative) to 5+ (strongest positive). For papain (NHSBT Reagents) treatment of RBCs, 1 volume of washed packed RBCs was incubated with 2 volumes of papain for 3 minutes, according to the manufacturer’s instructions. Following incubation, RBCs were washed a minimum of 4 times with phosphate buffered saline (PBS) until wash supernatant was clear.
Next-generation sequencing library preparation
Genomic DNA (gDNA) was extracted using a DNA isolation kit according to the manufacturer’s instructions (QIAamp DNA Blood Mini Kit; Qiagen). Exome sequencing was carried out using Nextera Rapid Capture Exome kit (Illumina), following the manufacturer’s instructions. Paired-end (2 × 87 cycle) single-plex sequencing was carried out on a MiSeq (Illumina). Secondary data analysis, including alignment of reads against human reference genome hg19, was carried out using MiSeq Reporter v2.5.1 (Illumina). Variants were called and annotated using Illumina BaseSpace Variant Interpreter. Homozygous or compound heterozygous missense variations with allele frequency <1% were considered as potential causes of loss of Er high-incidence antigen expression. Selected alignments were visualized using Integrative Genomics Viewer v2.8.13.27
Genetic analysis of PIEZO1
To confirm mutations identified using next-generation sequencing, polymerase chain reaction (PCR) primers for amplification of 51 PIEZO1 exons were designed using a primer designing tool (Primer-BLAST), with Homo sapiens chromosome 16, GRCh38.p13 Primary Assembly (NC_000016.10) as reference. Primer sequences and PCR conditions are shown in supplemental Table 2. Sanger sequencing of PCR products was carried out with forward and reverse PCR primers using a capillary automated DNA sequencer (3130xL Genetic Analyser; Applied Biosystems). Exon sequences were aligned to PIEZO1 reference using SeqScape software v.3 (Applied Biosystems).
Lentiviral transduction of Bristol erythroid line–adult (BEL-A) cells
For vector preparation, HEK293T cells (Clontech) were cultured in Dulbecco's Modified Eagle Medium containing 10% fetal calf serum (Gibco). Cells were seeded in 10 cm dishes, and calcium phosphate was transfected using lentiviral packaging vectors pMD2 (5 μg) and pPAX (15 μg) and plentiCRISPRv2 PIEZO1 gRNA (CATCCCCAACGCCATCCGGC) (Genscript) or XLG3 for expression of PIEZO1 open reading frames (20 μg). PIEZO1 gene synthesis and site-directed mutagenesis was performed by Genscript. After 24 hours, Dulbecco's Modified Eagle Medium was removed and replaced with 5 mL fresh media. The virus was harvested after 48 hours, concentrated using the Lenti-X concentrator (Clontech) and stored at −80°C.
Fluorescence-activated cell sorting (FACS) based on Piezo1 function
Piezo1 function was assessed using Yoda1, a small molecule Piezo1 selective agonist that induces Piezo1-mediated influx of calcium ions.30 Pretreatment of cells with calcium-responsive fluorescent dye FLUO4, before a challenge with Yoda1, enables detection of functional Piezo1 as assayed by an increase in fluorescent intensity of FLUO4, indicative of calcium influx via this channel. BEL-A cells transduced with plentiCRISPRv2 containing a guide targeting PIEZO1 exon 5, were loaded with 5 μM FLUO4 in Stemspan SFEM (StemCell Technologies) containing expansion growth factors for 30 minutes at 37°C. Cells were pelleted and resuspended in Iscove's Modified Dulbecco's Medium containing 2% fetal calf serum for FACS sorting. Immediately before FACS, Yoda1 (Sigma) was added to a final concentration of 20 μM for 1 minute and cells were subsequently sorted or screened on the basis of their response to the Yoda1 stimulus. Cells were FACS sorted into a positively responsive population, where Piezo1 was exogenously expressed. For derivation of PIEZO1 knockout (KO) clones, CRISPR-targeted cells were blind sorted into individual clones and functionally screened for KO on the basis of the lack of Yoda1 response.
Flow cytometry
For flow cytometry, 7.5 × 104 BEL-A cells resuspended in PBSAG (PBS + 1 mg/mL BSA, 2 mg/mL glucose) + 1% BSA were labeled with primary human antibody eluates at indicated dilutions for 30 minutes at 4°C. Cells were washed in PBSAG, incubated for 30 minutes at 4°C with Alexa647-conjugated antihuman secondary antibody, and washed. Data were acquired on a MacsQuant VYB Analyser (Miltenyi Biotec). Propidium iodide was used for exclusion of dead cells. For Yoda1 screening of transduced and sorted populations, methodology as described for FACS was employed.
RBC membrane preparation and immunoprecipitation
RBC membrane preparation and immunoprecipitation were performed as previously described.31 Immune precipitates were prepared from test Er(a+b–) donor RBCs and negative control Er(a–b+) (P1) RBCs using anti-Era plasma from P3 (5:1 ratio of plasma to cells). Proteomic analysis was performed at the University of Bristol Proteomics Facility (detailed in supplemental Methods, available on the Blood website).
Automated patch clamp analysis
Piezo1 channel activity in BEL-A cells overexpressing each individual Piezo1 variant (Glu2392Lys, Gly2394Ser, Glu2407Gln, Glu2407Lys, Arg2245Gln) and wild-type Piezo1 was assessed using a 384-well planar patch clamp (SyncroPatch 384PE; Nanion Technologies) as previously described32 with modifications detailed in supplemental Methods (available on the Blood website).
Homology modeling and molecular dynamics calculations
A homology model of amino acids 570 to 2521 was created using mouse Piezo1 homolog experimental coordinates derived from cryo-EM (PDB 5Z10)8 with human Piezo1 as a threading sequence using automated SWISS-MODEL server,33 resulting in an overall sequence identity of 82% for this region. A combined lipid-protein model was generated and analyzed as described in detail in supplemental Methods (available on the Blood website).
Results
Serologic characterization of Er alloimmunized individuals
Comprehensive serologic cross-compatibility testing was carried out with samples from known Er(a–b+), Er(a+b+) and Er(a–b–) individuals, plus the additional 3 individuals (and family members) included in this study due to variant Er expression and/or identification of apparently Er-related antibodies in their serum (Table 1; supplemental Table 1, available on the Blood website). Cross-compatibility testing (Table 2) clearly shows the expected reactivity of anti-Era, anti-Erb and anti-Er3 with Er(a–b+), Er(a+b+) and Er(a–b–) red cells. The antibodies made in unrelated individuals P11 and P12 were incompatible with Er(a–b+) and Er(a–b–) cells, but their red cells were mutually compatible, and compatible with cells from 4 siblings of P12. This indicates that P11 and P12 antibodies define the same novel antigen, designated Er4 in this study, and these individuals (and 4 compatible siblings) have the Er4– phenotype. The antibody present in the serum of P13 was found to be incompatible with Er(a–b+), Er(a–b–), and Er4– cells. This antibody defines a different novel antigen, designated Er5 in this study, and the red cells of P13 are Er5–.
Determination of the genetic basis of Era and Erb antigens
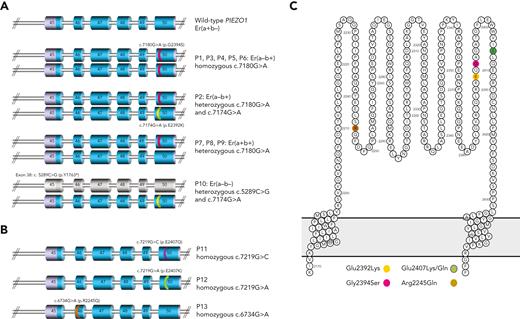
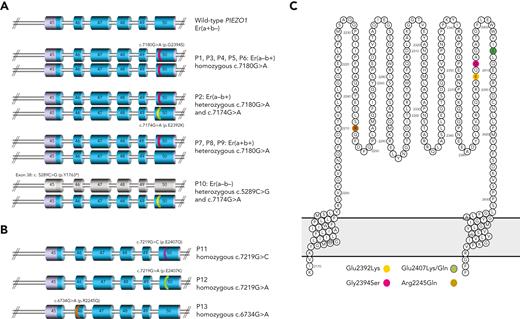
To uncover the genetic basis of the Er antigens, DNA from 2 unrelated Er(a–b+) individuals (P1, P2) was subjected to whole exome sequence analysis. Initial data filtering was used to search for genes carrying rare (frequency <0.01), homozygous missense mutations, consistent with observed lack of the high incidence Era antigen in both individuals. No variant fitting these criteria was shared by both individuals (supplemental Table 3, available on the Blood website); however, 1 rare homozygous missense PIEZO1 variant in P1 was found to be heterozygous in P2, so PIEZO1 was selected for further analysis. P1 was homozygous for 7180G>A (Gly2394Ser), in exon 50 of PIEZO1 (rs201950081; gnomAD freq. 0.0012), while P2 carried this same mutation in compound heterozygosity with a second nearby mutation, 7174G>A (Glu2392Lys; rs528448732; gnomAD freq. 0.0004). Mutations were confirmed by Sanger sequencing, and a further 4 unrelated Er(a–b+) samples (P3-P6) were also homozygous for 7180G>A. Three Er(a+b+) samples (P7-P9) were heterozygous for this mutation, consistent with 7180G>A encoding the Era/Erb polymorphism (7180G; Gly2394 associated with Era, 7180A; Ser2394 associated with Erb) (Figure 2A; supplemental Table 4, available on the Blood website).
Mutations in PIEZO1 gene (exons 45-50) encoding amino acid substitutions in Piezo1 protein extracellular loop. (A) PIEZO1 genotypes observed in Er(a−b+) and Er(a−b−) individuals. PIEZO1 exons 45 to 50 are shown (regions coding for extracellular domain shown in blue). Mutations are highlighted in pink (7180G>A) and yellow (7174G>A), seen in homozygous, heterozygous, and compound heterozygous states in individuals tested as shown. Individual P10 has nonsense mutation in exon 38 (not shown) predicted to encode truncated protein (exons colored gray). (B) PIEZO1 alleles observed in Er variant individuals. PIEZO1 exons 45 to 50 are shown. Mutations are highlighted in purple (7219G>C), green (7219G>A), and brown (6734G>A). All mutations were homozygous in the indicated individuals. (C) All mutations shown in panels A and B encode amino acid substitutions in the extracellular domain of Piezo1 protein (amino acids 2198-2431) as shown (colors as in 2A and 2B).
Mutations in PIEZO1 gene (exons 45-50) encoding amino acid substitutions in Piezo1 protein extracellular loop. (A) PIEZO1 genotypes observed in Er(a−b+) and Er(a−b−) individuals. PIEZO1 exons 45 to 50 are shown (regions coding for extracellular domain shown in blue). Mutations are highlighted in pink (7180G>A) and yellow (7174G>A), seen in homozygous, heterozygous, and compound heterozygous states in individuals tested as shown. Individual P10 has nonsense mutation in exon 38 (not shown) predicted to encode truncated protein (exons colored gray). (B) PIEZO1 alleles observed in Er variant individuals. PIEZO1 exons 45 to 50 are shown. Mutations are highlighted in purple (7219G>C), green (7219G>A), and brown (6734G>A). All mutations were homozygous in the indicated individuals. (C) All mutations shown in panels A and B encode amino acid substitutions in the extracellular domain of Piezo1 protein (amino acids 2198-2431) as shown (colors as in 2A and 2B).
P10, an Er(a–b–) individual, was shown to be compound heterozygous for 7174G>A (Glu2392Lys) and a nonsense mutation in exon 38 of PIEZO1 (5289C>G; Tyr1763Ter; rs72811487; gnomAD freq. 0.0001). The 7174G>A mutation, also observed in P2, appears to prevent expression of Era antigen when carried in cis with wild-type 7180G (Figure 2A; supplemental Table 4, available on the Blood website).
Genetic investigation of families with suspected Er variant phenotype
Three further individuals with suspected Er-related antibodies, as well as their respective family members (supplemental Table 1, available on the Blood website), were also subjected to Sanger or whole exome sequencing of PIEZO1. Individual P11, with predicted Er variant phenotype (Er4–), was found to carry a rare homozygous missense mutation (7219G>C; Glu2407Gln; rs200291894; gnomAD freq. 0.0001) in exon 50 of PIEZO1, while her children (P11-F1 and P11-F2) were heterozygous for this mutation (Figure 2B; supplemental Table 4, available on the Blood website).
A similar rare homozygous missense mutation, affecting the same PIEZO1 nucleotide but resulting in a different amino acid substitution, was observed in P12 (7219G>A; Glu2407Lys; rs200291894; gnomAD freq. 0.0001). This mutation was demonstrated to segregate in 11 tested family members (four 7219A homozygotes were serologically compatible while 5 heterozygotes and 2 wild-type 7219G were incompatible; supplemental Figure 1, available on the Blood website).
One further mutation was identified in PIEZO1 exon 46 (6734G>A; Arg2245Gln; rs2290901; gnomAD freq. 0.0033) as the potential causative mutation in an HDFN case, carried in the homozygous state in the alloimmunized mother P13 (Er5–) and heterozygous state in her child (while the father showed wild-type sequence at this position).
All PIEZO1 mutations identified by exome/Sanger sequencing are detailed in Figure 2 and supplemental Table 4 (available on the Blood website). As PIEZO1 encodes a known transmembrane protein, expressed on erythrocytes, with all observed mutations predicted to encode amino acid substitutions falling within the extracellular region of the protein (amino acids 2198-2431; Figure 2C), this was considered a good candidate for the carrier of the Er blood group antigens.
Disruption of PIEZO1 ablates Er antibody binding to erythroid cells
To create a cellular model for establishing specificity of anti-Er alloantibodies for Piezo1, CRISPR-mediated gene editing was applied to BEL-A erythroblast cell line, generating a novel human erythroid PIEZO1 KO cell line. Expanding BEL-A cells were transduced with a lentiviral vector expressing Cas9 and a guide targeting PIEZO1 exon 5. Because endogenous surface Piezo1 expression was not detectable with commercially available antibodies, a functional assay using FLUO4 and Yoda1 stimulus was used to screen for KO (Yoda1 unresponsive) clones (supplemental Figure 2A, available on the Blood website). Sequencing of a selected clone confirmed presence of a biallelic disruptive PIEZO1 mutation; 354_361delCATCCGGC (Ile119GlyfsTer4) (supplemental Figure 2B, available on the Blood website), confirming both genotypic and functional Piezo1 knockout.
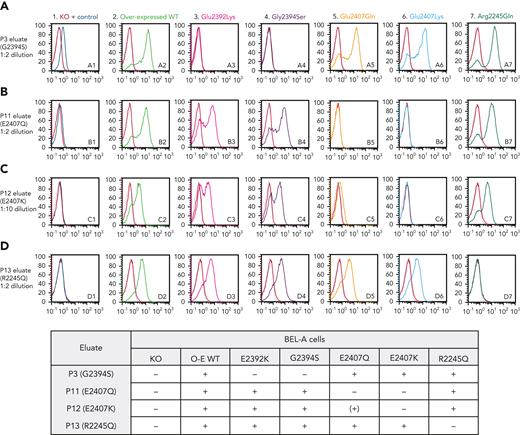
Flow cytometry using anti-Era, isolated from alloimmunized Er(a–b+) individual P3, was able to detect antibody binding to unmodified BEL-A cells that was completely ablated in PIEZO1 KO cells, confirming Piezo1 as the carrier molecule for the Era antigen (Figure 3, A1).
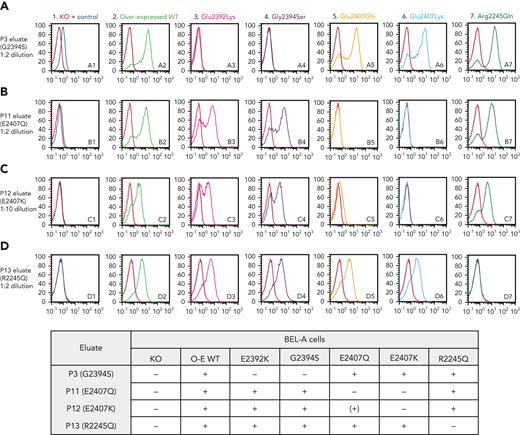
Anti-Er alloantibodies are specific for antigenic sites on Piezo1. Flow cytometry histograms illustrate cell surface labeling of indicated wild-type or mutant Piezo1 constructs overexpressed in an endogenous Piezo1 knockout (KO) BEL-A cell line using antibodies eluted from plasma of individuals (P3, row A; P11, row B; P12, row C; and P13, row D) with mutations in Piezo1 as labeled. BEL-A Piezo1 KO cells and endogenous Piezo1 are shown in column 1, with overexpressed wild-type Piezo1 in column 2. Columns 3 to 7 show overexpressed mutant Piezo1 constructs as labeled. Results are summarized in the grid below histograms. O-E WT, overexpressed wild-type; +, positive; −, negative; (+), weakly positive.
Anti-Er alloantibodies are specific for antigenic sites on Piezo1. Flow cytometry histograms illustrate cell surface labeling of indicated wild-type or mutant Piezo1 constructs overexpressed in an endogenous Piezo1 knockout (KO) BEL-A cell line using antibodies eluted from plasma of individuals (P3, row A; P11, row B; P12, row C; and P13, row D) with mutations in Piezo1 as labeled. BEL-A Piezo1 KO cells and endogenous Piezo1 are shown in column 1, with overexpressed wild-type Piezo1 in column 2. Columns 3 to 7 show overexpressed mutant Piezo1 constructs as labeled. Results are summarized in the grid below histograms. O-E WT, overexpressed wild-type; +, positive; −, negative; (+), weakly positive.
Expression of Piezo1 mutants confirms Er antigen loci
To determine specificity of antibodies eluted from sera of individuals carrying different PIEZO1 mutations, lentiviral constructs were generated overexpressing full-length wild-type Piezo1, as well as Piezo1 with 1 of the following mutations individually introduced: Glu2392Lys, Gly2394Ser, Glu2407Gln, Glu2407Lys, Arg2245Gln. Constructs were transduced into the endogenous Piezo1 KO BEL-A clone, and successfully transduced populations were selected by FACS sorting of Yoda1 responsive cells. In all cases, mutated Piezo1 retains Yoda1 responsivity (supplemental Figure 3, available on the Blood website).
Antibody eluates produced from 4 individuals carrying different PIEZO1 mutations—P3 (Gly2394Ser), P11 (Glu2407Gln), P12 (Glu2407Lys), and P13 (Arg2245Gln)—were tested by flow cytometry against unmodified and Piezo1 KO BEL-A cells, as well as against cells overexpressing wild-type and each mutant Piezo1 construct.
Only P3 and P11 eluates were able to weakly detect endogenous BEL-A Piezo1 (Figure 3, column 1); however, all eluates showed strong positive reactions with overexpressed wild-type Piezo1 (Figure 3, column 2), with specificity of antibody binding to Piezo1 demonstrated by absence of labeling of the KO clone in each case. Each tested eluate gave expected negative reactions with cells overexpressing Piezo1 harboring their corresponding mutation.
P3 eluate (anti-Era, Figure 3, row A) reacted strongly with cells overexpressing Piezo1 harboring either Glu2407Gln, Glu2407Lys, or Arg2245Gln mutation, while negative reactions were observed with Gly2394Ser and Glu2392Lys. This pattern of reactivity was replicated using a further example of anti-Era from P2 (data not shown). Anti-Era reacted with all Piezo1 constructs expressing Gly2394, demonstrating that this residue is required for Era expression, while Ser2394 presumably defines the antithetical Erb antigen. The Glu2392Lys mutation appears to result in absence of expression of Era (despite the presence of Gly2394), as demonstrated by lack of reactivity with either anti-Era eluate (P3 or P2).
P11 and P12 eluates (Figure 3, rows B and C) both reacted strongly with Piezo1 harboring Glu2392Lys or Gly2394Ser, as well as with Arg2245Gln mutant cells, demonstrating that these antibodies recognize a different antigen to Era, here defined as Er4. P11 and P12 both have mutations affecting the same amino acid residue of Piezo1 (Glu2407Gln and Glu2407Lys, respectively). These individuals appear serologically compatible, and P11 eluate failed to react with Glu2407Lys mutant cells. Interestingly, however, P12 eluate showed a markedly reduced, but detectable, reactivity with Glu2407Gln Piezo1 (Figure 3, C5). This likely reflects reduced disruption to the epitope brought about by a Glu>Gln mutation compared with the Glu>Lys mutation in P12.
P13 eluate (Figure 3, row D) robustly detected each of the other mutated Piezo1 proteins (Glu2392Lys, Gly2394Ser, Glu2407Gln, Glu2407Lys), showing that it recognizes a further different antigen from Era or the proposed Er4 antigen, defined here as Er5.
Similar results were obtained from overexpression studies in a second Piezo1 KO BEL-A clone (supplemental Figure 4, available on the Blood website).
Immunoprecipitation using anti-Era
Further evidence for Piezo1 carrying the Er antigens was obtained by immunoprecipitation of red cell membranes prepared from test Er(a+b–) RBCs and negative control Er(a–b+) RBCs, using human anti-Era (from P3). Proteomic analysis, comparing test immunoprecipitation to negative control, showed Piezo1 to be the most overrepresented protein in the test immunoprecipitation by a considerable margin (score ratio test: negative >500; supplemental Table 5, available on the Blood website). These data indicate that anti-Era can selectively bind Piezo1 in Er(a+) cells, but not in Er(a–) RBCs.
Functional characterization of Piezo1 mutants using high-throughput automated patch clamp
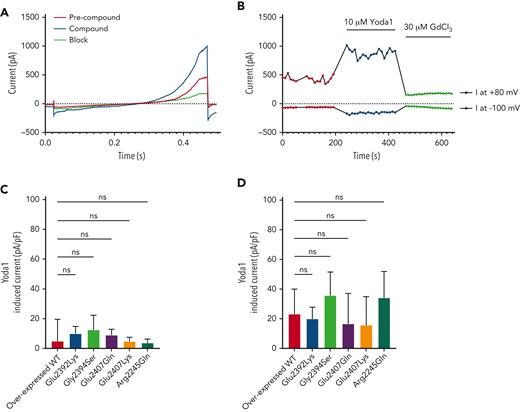
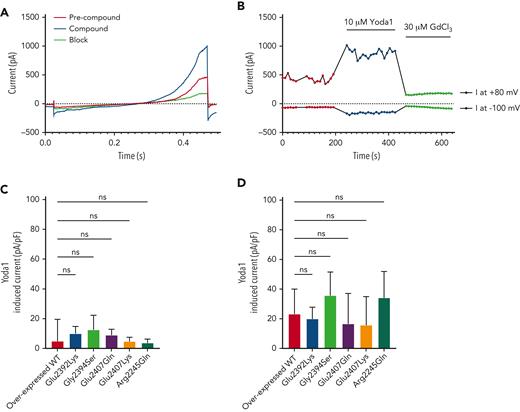
To investigate potential functional effects, Piezo1 current responses from mutant Piezo1 constructs overexpressed in BEL-A cells were compared with wild-type Piezo1 constructs. Whole cell currents were recorded in response to a voltage ramp protocol (−100 mV to 80 mV, Figure 4A), which elicited an outward rectifying current (Figure 4A-B red trace/symbols). Yoda1 was applied to enhance activity, triggering current amplitude increases at both −100 mV and 80 mV, indicative of nonselective cation channels such as Piezo1 (Figure 4A-B, blue trace/symbols). Subsequent addition of GdCl3, a nonspecific blocker of mechanosensitive channels, blocked the current (Figure 4A-B, green trace/symbols) as previously described.32 Current amplitudes in response to Yoda1 showed no significant difference in magnitude (Figure 4C) or percentage of responding cells (supplemental Figure 5A, available on the Blood website) between mutants and wild types, indicating normal Piezo1 functionality for all mutants. To further isolate Piezo1 currents, experiments were repeated in the presence of TRAM-34, a specific Gardos channel blocker. Yoda1-induced currents were amplified but also exhibited no significant differences between mutant and wild-type Piezo1 (Figure 4D; supplemental Figure 5B, available on the Blood website).
Evaluation of Piezo1 activity in BEL-A overexpressed wild-type and mutant cells using patch clamp. Raw traces of current amplitudes (A) and time course of the experiment (B) at 80 and −100 mV from an example overexpressed wild-type (WT) Piezo1 cell obtained before (precompound, red trace/symbols) and after (compound, blue trace/symbols) addition of 10 μM Yoda1, using a voltage ramp protocol (−100 to 80 mV, 400 ms, holding potential −30 mV) and inhibited by 30 μM GdCl3 (block, green trace/symbols). (C) Evaluation of current increase on Yoda1 addition (Yoda1 induced current, IYoda1− IES, pA/pF) of responding/nonresponding cells identified from BEL-A overexpressed wild-type and mutant Piezo1 constructs (n = 11/77 overexpressed WT; n = 21/84 Glu2392Lys; n = 13/80 Gly2394Ser; n = 15/89 Glu2407Gln; n = 15/80 Glu2407Lys; n = 19/85 Arg2245Gln). (D) Evaluation of current increase on Yoda1 addition in the presence of TRAM-34 (Yoda1 + TRAM-34 induced current, IYoda1− IES, pA/pF) of responding cells identified from BEL-A overexpressed wild-type and mutant Piezo1 constructs (n = 48/82 overexpressed WT; n = 54/86 Glu2392Lys; n = 62/87 Gly2394Ser; n = 47/93 Glu2407Gln; n = 47/77 Glu2407Lys; n = 45/84 Arg2245Gln).
Evaluation of Piezo1 activity in BEL-A overexpressed wild-type and mutant cells using patch clamp. Raw traces of current amplitudes (A) and time course of the experiment (B) at 80 and −100 mV from an example overexpressed wild-type (WT) Piezo1 cell obtained before (precompound, red trace/symbols) and after (compound, blue trace/symbols) addition of 10 μM Yoda1, using a voltage ramp protocol (−100 to 80 mV, 400 ms, holding potential −30 mV) and inhibited by 30 μM GdCl3 (block, green trace/symbols). (C) Evaluation of current increase on Yoda1 addition (Yoda1 induced current, IYoda1− IES, pA/pF) of responding/nonresponding cells identified from BEL-A overexpressed wild-type and mutant Piezo1 constructs (n = 11/77 overexpressed WT; n = 21/84 Glu2392Lys; n = 13/80 Gly2394Ser; n = 15/89 Glu2407Gln; n = 15/80 Glu2407Lys; n = 19/85 Arg2245Gln). (D) Evaluation of current increase on Yoda1 addition in the presence of TRAM-34 (Yoda1 + TRAM-34 induced current, IYoda1− IES, pA/pF) of responding cells identified from BEL-A overexpressed wild-type and mutant Piezo1 constructs (n = 48/82 overexpressed WT; n = 54/86 Glu2392Lys; n = 62/87 Gly2394Ser; n = 47/93 Glu2407Gln; n = 47/77 Glu2407Lys; n = 45/84 Arg2245Gln).
Homology modeling of Er antigenic sites on Piezo1 protein
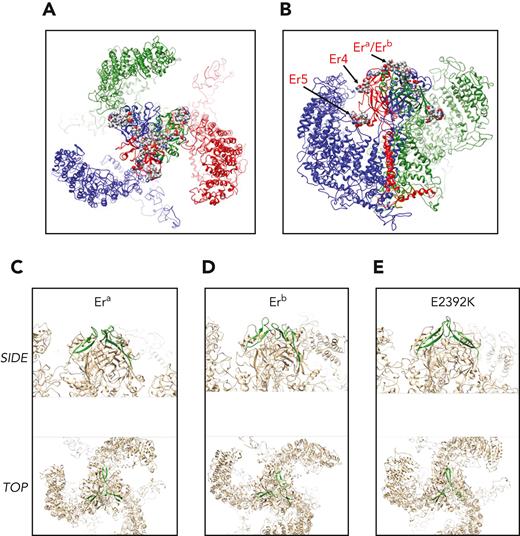
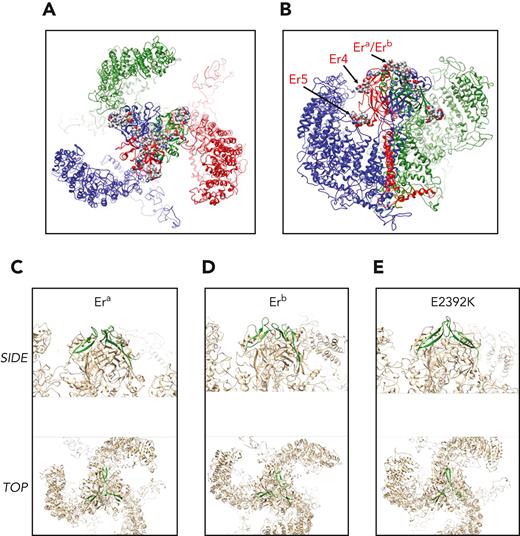
Molecular models of wild-type and mutant Piezo1 were analyzed by molecular dynamic calculations. Wild-type Piezo1 and an overview of the locations of Er antigenic determinants are shown in Figure 5. Era and Erb are contained in an apical loop extending from the center of the trimer (Figure 5A-B). To assess potential impacts of substitutions, the Era (Gly2394; Figure 5C), Erb (Ser2394; Figure 5D), and Glu2392Lys (Figure 5E) substitutions were analyzed using implicit solvent models (as described in supplemental Methods, available on the Blood website). A trajectory of 100 ns calculation time was subsequently analyzed for potential impact of substitutions on presentation of combined antigenic sites. Clustering restricted to the loop structure presented by amino acids 2384 to 2408 (so as not to bias clustering results toward dominating conformers in distal regions of the protein complex) was compared for the 3 trajectories. The highest populated cluster is highlighted for comparison in Figure 5C-E. The very apical section of the trimer extending to the extracellular side is made up entirely from the combined sites of the 3 exposed loops encompassing amino acids E2392QGAGATG2399. Er4 (Glu2407Lys) is located at the end of a beta sheet connecting to the Era/Erb antigenic loop region. Er5 (Arg2245Gln) is in a short ɑ-helical segment at the membrane-proximal side of the extracellular trimeric domain (as indicated in Figure 5B).
Modeling of Piezo1 protein and location of Er antigenic sites. A homology model for the human Piezo1 sequence was generated using coordinates form PDB ID 5Z1016 and inserted into a model membrane for subsequent molecular dynamics calculations for a total trajectory time of 100 ns. (A) Top view on extracellular side of equilibrated Piezo1 trimer (composed of 3 chains: red, green, and blue). Lipids have been omitted from the illustration for simplicity. The residues encompassing the antigenic sites are shown as spheres (colored by atom type) located in and around the center of the trimer. (B) Side-view slice of the Piezo1 trimer with the transmembrane section of 1 monomer (red) removed to allow view on center. Representation is the same as in panel A. Side chains of antigenic sites (Era/Erb, Er4, and Er5) are highlighted for the red monomer. (C-E) Era/Erb loops as predicted by molecular dynamics calculations. The backbones of loops encompassing residues 2384 to 2408 are shown in green with the remainder of the protein trimer in beige. The conformation (in green) is representative of the highest populated cluster in the trajectory. The first row represents a side view onto the central extracellular domain, and below it is a depiction of the apical view on the membrane plane as indicated in figure panels. (C) Era representative loop. (D) Erb representative loop. (E) Glu2392Lys representative loop.
Modeling of Piezo1 protein and location of Er antigenic sites. A homology model for the human Piezo1 sequence was generated using coordinates form PDB ID 5Z1016 and inserted into a model membrane for subsequent molecular dynamics calculations for a total trajectory time of 100 ns. (A) Top view on extracellular side of equilibrated Piezo1 trimer (composed of 3 chains: red, green, and blue). Lipids have been omitted from the illustration for simplicity. The residues encompassing the antigenic sites are shown as spheres (colored by atom type) located in and around the center of the trimer. (B) Side-view slice of the Piezo1 trimer with the transmembrane section of 1 monomer (red) removed to allow view on center. Representation is the same as in panel A. Side chains of antigenic sites (Era/Erb, Er4, and Er5) are highlighted for the red monomer. (C-E) Era/Erb loops as predicted by molecular dynamics calculations. The backbones of loops encompassing residues 2384 to 2408 are shown in green with the remainder of the protein trimer in beige. The conformation (in green) is representative of the highest populated cluster in the trajectory. The first row represents a side view onto the central extracellular domain, and below it is a depiction of the apical view on the membrane plane as indicated in figure panels. (C) Era representative loop. (D) Erb representative loop. (E) Glu2392Lys representative loop.
The loop conformations encompassing Era/Erb were observed to give rise to considerably divergent loop structures (supplemental Figure 6, available on the Blood website). Era was observed to exhibit similar conformers in the most highly populated clusters across the trajectory, whereas Erb was seen to be populated by more divergent conformers. Substitution of Glu2392 by lysine may lead to relatively divergent loop formations relative to Era/Erb. This may be due to potential reshuffling of salt bridges in this apical region of the trimer. In particular, formation of a new salt bridge between Lys2392 and Glu2402, as well as Glu2410, was observed, replacing existing salt bridges for the 2 glutamic acid residues in the wild-type trajectory (supplemental Figure 7, available on the Blood website).
Discussion
Nearly 40 years have passed since the high-incidence red blood cell Era antigen was first described, and throughout this period its molecular basis has remained elusive. In this study we demonstrate the mechanosensitive ion channel Piezo1 as the site of 3 red blood cell antigens—Era, Erb, and Er3—and further identify 2 novel high-incidence antigens, described here as Er4 and Er5. Our findings show that Gly2394 is required for expression of the high-incidence Era antigen, while Ser2394 encodes the antithetical low-incidence antigen, Erb. Modeling shows this amino acid residue to be on an exposed apical protein loop and reveals significant predicted differences in structure between Gly2394 and Ser2394. Proposed novel high-incidence antigens Er4 and Er5 are associated with Glu2407 and Arg2245 in Piezo1, respectively. Modeling is consistent with these residues being antigenic, with the Er4 antigenic site adjacent to the Era/Erb loop region, and Er5 located in a membrane-proximal extracellular domain. Our data support the elevation of Er from blood group collection to blood group system status.
Lack of the high-incidence Er3 antigen does not appear to be associated with a true null phenotype as originally proposed4 but rather represents lack of both Era and Erb antigen expression. The Er(a–b–), Er3–, individual (P10) genetically characterized in this study expresses a variant Piezo1 protein, carrying both Er4 and Er5. Although 1 allele is predicted to encode a truncated protein, lacking the external loop region, and consequently expressing no Er antigens, the other allele in this patient encodes wild-type Gly2394 (Era) with a nearby Glu2392Lys mutation. Modeling suggests this mutation causes a significant conformational change around the Era epitope, resulting in the serologically observed lack of Era expression (without altering expression of Er4 or Er5). There are likely to be other genetic backgrounds resulting in Er(a–b–) phenotypes, and the specificity of antibodies historically known as anti-Er3 may be heterogeneous. It is unclear whether complete lack of Piezo1 is compatible with life, as although mice deficient in Piezo1 die in utero,12 patients with homozygous PIEZO1 nonsense mutations have survived to adulthood.15 As these would be predicted to result in truncated proteins lacking the Er antigen domain, the study of such patients could greatly aid characterization of Er antigen expression and the role of Er antibodies in transfusion.
PIEZO1 is a highly polymorphic gene, implicated in multiple diseases,13,15-17,20,22,23 with a host of additional nonpathogenic SNPs reported. A recent study estimated that as many as 1 in 8000 Americans may present symptoms of DHS.34 DHS most commonly results from heterozygous pathogenic gain of function mutations in PIEZO1, although extremely rare homozygous mutations have also been reported.16,35 Interestingly, several studies using targeted next-generation sequencing hereditary anemia gene panels have associated Gly2394Ser, identified here as the antigenic site of Era/Erb, with DHS.36-38 However, there is no suggestion in the Er literature that any RBC pathology or morphologic abnormalities are associated with the rare Er(a–b+) phenotype. All Er(a–b+) and Er(a+b+) samples in this study were archive material, and thus no data are available to ascertain any potential functional significance of this mutation.
The Glu2407Gln mutation, associated with Er4 antigen expression in this study, has also been reported to be pathogenic,38 and the potential functional significance of the other variants identified (Glu2407Lys; Er4 and Arg2245Gln; Er5) requires further investigation. Unfortunately, fresh patient blood samples were unavailable to perform ektacytometry and obtaining relevant clinical data was challenging. Data from patients P12 (and family) and P13 are included in supplemental Tables 6 and 7 (available on the Blood website; P11 data unavailable as patient is deceased). Interpretation is complicated by the existence of additional underlying conditions in both P12 and P13. P12 homozygous variant family members show no signs of macrocytosis or reticulocytosis, but there is a tendency toward increased mean cell hemoglobin concentration, possibly indicating some degree of red cell dehydration. P13 red cells were hypochromic and microcytic, consistent with a coincident ɑ-thalassemia trait. Blood film examination of both P12 and P13 showed no evidence of stomatocytes, and no clinical or laboratory features of hemolysis were observed. No electrophysiologic abnormalities were detected in BEL-A cells overexpressing any of the Piezo1 variants as compared with wild-type, suggesting that these variants, although antigenic, may not be of particular relevance for Piezo1 function. However, an associated DHS phenotype cannot be conclusively excluded, and the possibility of an overlap between Er antigen expression and red cell morphologic abnormalities warrants further investigation.
The Gly2394Ser, Glu2407Gln, and Glu2407Lys mutations are rare in all populations (supplemental Table 8, available on the Blood website), with the highest frequency being that of Gly2394Ser in the European population (freq. 0.002). Homozygosity for these variants, as required for loss of associated high-incidence Er antigens, is predicted to be extremely rare, as supported by scarcity of reported alloimmunization events. Interestingly, Arg2245Gln, although rare (freq. 0.003), is significantly overrepresented in African populations (freq. 0.03), so the Er5–phenotype (and associated alloimmunization events) will be more frequent in patients of African descent. This observation may also point to a wider functional effect of this mutation. Although there have been conflicting reports,39 studies report that a gain-of-function PIEZO1 mutation, E756del, common in African populations, causes red cell dehydration and/or is protective against malaria.40,41 Similar protective variants may be under positive selection in African populations (and thus overrepresented), so the role of the Arg2245Gln variant requires further study.
As reported cases of Er alloimmunization are rare, there remains sparse evidence to conclusively establish or exclude a role of such antibodies in HTR or HDFN. The majority of Er antibodies in this study were identified during pregnancy, with 2 patients presenting with apparent severe HDFN, resulting in fetal/neonatal death (P12; Er4 and P13; Er5; supplemental Table 1, available on the Blood website). Interestingly, P12 had 5 uneventful pregnancies, only experiencing complications in her sixth pregnancy, following a potentially immunizing transfusion event 2 years previously. Therefore, anti-Er4 and anti-Er5 appear clinically significant, although the reported association of PIEZO1 mutations with nonimmune hydrops fetalis15,23 adds complication to unpicking any role of Er antibodies in HDFN.
Considering the high level of polymorphism of PIEZO1, including nearly 200 missense mutations reported within the antigenic extracellular region of the protein, it seems likely that further antigens, of both high and low incidence, will be added to the Er system in the future. Further investigation of the amino acid changes identified in P11 and P12 may be warranted because, although these individuals are serologically compatible, the flow cytometry data showed an intriguing hint of a difference in antibody reactivity.
In a relatively short period of time after its discovery, understanding of the structure, mechanism, and function of Piezo1 has burgeoned, with important functions established within red cell morphology, erythropoiesis, reticulocyte maturation, and susceptibility to malaria infection. Demonstration of Piezo1 as the carrier molecule for the long recognized Er red cell antigens establishes a new blood group system and further highlights the importance of this protein. Piezo1 is present at only a few hundred copies on the surface of the red cell,26 reminding us that low copy number is a barrier to neither functional importance nor antigenicity. The occurrence of fatal HDFN associated with antibodies directed against Er antigens highlights their clinical relevance, elucidating another facet of the importance of Piezo1 in human biology.
Acknowledgments
The authors thank the following individuals: Jan Frayne, Dave Anstee, and Kongtana Trakarnsanga for provision of the BEL-A cell line; Kate Heesom from the University of Bristol Proteomics Facility for mass spectrometry analysis of immune precipitates; Andrew Herman from the University of Bristol Faculty of Biomedical Sciences Flow Cytometry Facility for cell sorting support; IBGRL Molecular Diagnostics Department for Sanger sequencing support; Christian Jannete Stevens, from Bristol Institute of Transfusion Sciences, UK, for providing PIEZO1 amplification primers for exons 23, 24, and 39; and Geoff Daniels for critical review of the manuscript. The authors would also like to acknowledge the invaluable insights, advice, and support from Dave Anstee (1946-2021).
This study was supported in part by the National Institute for Health Research Blood and Transplant Research Unit (NIHR BTRU) in Red Cell Products (IS-BTU-1214-10032), NHSBT R&D funding (WP15-04 and WP-15-05), the Medical Research Council (MR/V010506/1), the Cultural Bureau at the Royal Saudi Embassy in the United Kingdom and the Saudi Ministry of Education (SAA PhD scholarship), and the European Union ITN “EVIDENCE” grant agreement ID 860436)
The views expressed are those of the authors and not necessarily those of the National Health Service, NIHR, or Department of Health and Social Care.
Authorship
Contribution: N.M.T., V.K.C., L.A.T., T.J.S., and A.M.T. conceived, designed, and coordinated the study; N.M.T. and B.J. performed serology experiments; L.A.T., V.K.C., and S.A.A. performed whole exome sequencing, Sanger sequencing, and associated data analysis; T.J.S. designed and performed CRISPR/Cas9 experiments (knockout and overexpression of Piezo1) and performed the flow cytometry experiments; P.J.W. carried out protein modeling and molecular dynamics calculations; F.A.S. and S.A.A. designed and carried out immunoprecipitation experiments; C.M.F., N.M., and M.G.R. conducted patch clamping optimization, experiments, and analysis under the supervision of N.B. and M.R.; S.J.W. detected and provided P11, P11-F1, and P11-F2 samples for antibody identification; M.H., R.A., and M.A. detected and referred samples P12 and P12-F1 to P12-F11 for antibody identification and provided clinical information; I.S., S.L., S.H., and J.J. detected and referred samples P13, P13-F1 and P13-F2 for antibody investigation; T.L., W.L., and M.D.K. provided comprehensive clinical data and interpretation for the case of P13; A.M.T. and N.M.T. provided advice on experimental design, analysis and interpretation of data, manuscript preparation, and critical review; N.M.T., V.K.C., L.A.T., S.A.A., and T.J.S. wrote the manuscript; and all authors contributed to review of the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicole M. Thornton, International Blood Group Reference Laboratory, NHS Blood and Transplant, 500 North Bristol Park, Northway, Filton, Bristol BS34 7QH, United Kingdom; e-mail: nicole.thornton@nhsbt.nhs.uk.
References
Author notes
Nucleotide sequence data are available in the DDBJ/EMBL/GenBank databases under the accession numbers OM914592, OM914593, OM914594, OM914595, OM914596, and OM914597. Mass spectrometry proteomics data are available from the PRIDE repository via ProteomeXchange with identifier PXD035013. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
V.K.C., L.A.T., and T.J.S. are joint first authors.