Introduction: Cyclin D1 protein expression in lymphocytes is commonly associated with mantle cell lymphoma. However, it has rarely been reported in patients with diffuse large B-cell leukemia (DLBCL), particularly in Asia where studies on cyclin D1 expression in DLBCL are limited and often small. Furthermore, the clinical implications of cyclin D1 expression in Asia remain unclear.
Methods: In this study, a total of 815 patients diagnosed with DLBCL and treated with R-CHOP or R-CHOP-like regimens over a five-year period were reviewed. The authors conducted clinical, immunohistochemical, and genetic analyses, with a specific focus on cyclin D1. Immunohistochemistry was used to identify cyclin D1-positive patients, and fluorescence in situ hybridization was employed to assess cyclin D1 gene (CCND1) rearrangements. A comparison was made between DLBCL patients with and without cyclin D1 expression in terms of clinical, immunohistochemical, and genetic features.
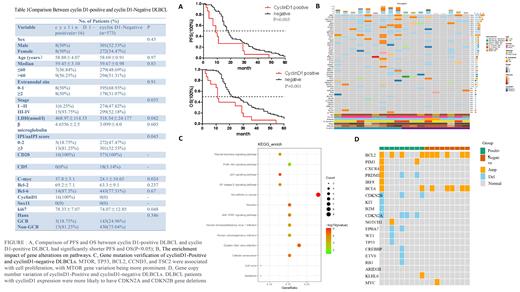
Results: Among the DLBCL patients, 16 individuals (2.61%) exhibited cyclin D1 expression without CCND1 gene rearrangements. Cyclin D1-positive DLBCLs were observed in patients with a median age of 58.88 ± 4.07 years, with 93.75% of them classified as stage IV. The majority of cyclin D1-positive patients (81.25%) displayed elevated serum lactate dehydrogenase (LDH) levels and advanced international prognostic index (IPI) scores. Additionally, all cases exhibited a proliferation rate exceeding 80%, and a significant proportion (93.75%) demonstrated MYC expression. Cyclin D1-positive DLBCLs were associated with elevated LDH levels, higher IPI scores, advanced stage, increased Ki-67 proliferation, and MYC expression (p<0.05). However, no significant differences were observed in other clinical characteristics and immunohistochemical features. Patients with cyclin D1 expression experienced shorter progression-free survival (PFS) and overall survival (OS) compared to cyclin D1-negative patients (p<0.05). Next-generation sequencing (NGS) was performed to explore the molecular mechanisms underlying cyclin D1-positive DLBCL. Among the top 50 gene alterations identified, all samples exhibited BRD9 and NOTCH2NL gene mutations, while gene alterations in the PI3K/AKT signaling pathways were evident in cyclin D1-positive DLBCLs. Furthermore, a more pronounced variation in the MTOR gene was observed between cyclin D1-positive and negative patients. Notably, DLBCL patients with cyclin D1 expression frequently exhibited CDKN2A and CDKN2B gene deletions, known to regulate the cell cycle transition from G1 to S phase in tumors.
Conclusion: Cyclin D1-positive DLBCL patients exhibited aggressive clinical behavior and inferior survival outcomes compared to DLBCL patients without cyclin D1 expression. NGS studies revealed a potential association between cyclin D1 expression and abnormal cell cycle transition and AKT pathway activation. These findings support the existence of a diagnostically challenging “gray zone” of DLBCL associated with cyclin D1 expression.
Disclosures
No relevant conflicts of interest to declare.