Introduction
Immunoglobulin light chain (AL) amyloidosis is a rare disease involving the clonal proliferation of bone-marrow-residing plasma cell and resulting in overproduction of serum immunoglobulin free light chains that affects multiple organs. There are several available effective treatments including autologous stem cell transplant, bortezomib, anti-CD38 antibodies, and immunomodulatory drugs. However, because of the atypical symptoms and signs of this disease, diagnostic delays are still the major challenge resulting in poor prognosis. In recent years, machine learning (ML) models have been used to assist in early diagnosis. Therefore, to address this clinical unmet need, we aim to build ML algorithms from clinical data and assess their performance in differentiating AL amyloidosis from similar conditions.
Methods
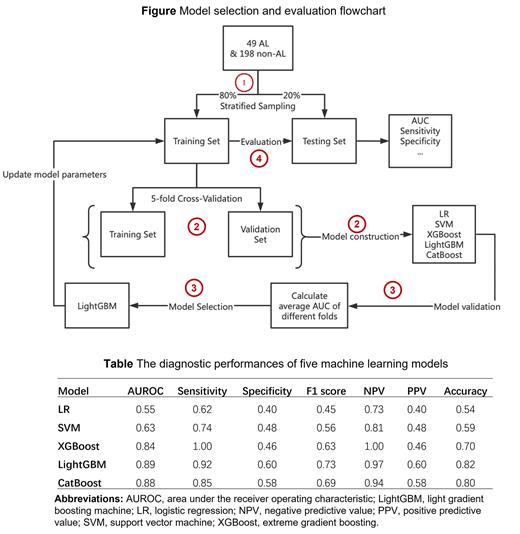
Monocenter medical records data were collected from 49 patients with AL amyloidosis and 198 non-AL amyloidosis patients on a ratio of 1:4 in Peking University People's Hospital between January 1, 2013, and December 31, 2021. The non-AL amyloidosis group were patients with diseases of similar symptoms including autoimmune liver disease, myocarditis, and hypertrophic cardiomyopathy. Variables for model development were selected from 30 demographic characteristics and clinical features from routine clinical examination based on the results of recursive feature elimination and hematologists' knowledge. We proposed a four-step approach to develop and evaluate the diagnostic models. In the first step, all patients were randomly allocated into a training set and a testing set with a ratio of 4:1. Second, we derived five separate ML models including logistic regression, support vector machine (SVM), extreme gradient boosting (XGBoost), light gradient boosting (LightGBM), and CatBoost algorithms to differentiate AL amyloidosis from other diseases with similar symptoms and validated the models using five-fold cross validation methods. Third, parameters of model with the highest areas under the receiver operating characteristic curves (AUROC) were updated in the full training set. Finally, the performances of the selected model were evaluated by AUROC, sensitivity, specificity and F1-score in the testing set.
Results
Twelve features including alanine aminotransferase, troponin, albumin, aspartate aminotransferase, activated partial thromboplastin time, albumin and globulin (A/G) ratio, direct bilirubin, platelet, fibrinogen, blood urea nitrogen, body weight and age were selected to construct ML models. The AUROC values for AL amyloidosis differential diagnosis were 0.55 with logistic regression, 0.63 with SVM, 0.84 with XGBoost, 0.89 with LightGBM, and 0.88 with CatBoost. The LightGBM model, which achieved the highest AUROC, also achieved the best performance with a sensitivity of 0.92, a specificity of 0.60, a F1-score of 0.73, a negative predictive value of 0.97, a positive predictive value of 0.60, and an accuracy of 0.82.
Conclusion
Our results show that the LightGBM model has the best performance to identify patients with AL amyloidosis from patients with similar symptoms. This novel ML-based diagnostic model has potential to assist in the earlier diagnosis of AL amyloidosis in clinical settings. Further studies are needed to confirm these findings in different study populations.
Disclosures
Lu:Jassen Pharmaceutical Ltd: Consultancy, Speakers Bureau.