Background:
AML is a life-threatening disease, and to determine which patients need allogeneic stem cell transplantation, hematologists risk-stratify each case. However, standard risk stratification using the European LeukemiaNet (ELN) criteria is focused on baseline mutations and chromosomal aberrations, and the risk estimate is not updated during a patient's course. In other blood cancers, recalculating the risk with treatment response data can help guide the need for more intensive therapy (Kurtz, et al, Cell, 2019). Furthermore, deep learning graph neural networks (GNN) applied to EHR data have strong predictive power in a hematology context (Fouladvand, et al, J Biomed Inform, 2023). Thus, we evaluated the power of a GNN to predict survival in AML using longitudinal EHR data, specifically with labs and histological features that are not included in the ELN but may capture the treatment response.
Methods:
Patients who were seen at the Stanford Cancer Institute, had EHR data available within six months of diagnosis, and were diagnosed with AML between June 1998 and January 2021 were included in this retrospective analysis. The GNN was trained to predict survival at two years from diagnosis using the first six months of clinical data. Patients were excluded if they were lost to follow-up before two years or died before six months. Data were collected from structured databases associated with Stanford's EHR, except that diagnosis dates were from Stanford's Cancer Registry, and survival data was supplemented with other databases including the Social Security Death Index. Dysplasia, bone marrow cellularity, and bone marrow blast percentages from pathology reports (“pathology report data”) were extracted using text processing algorithms and weakly supervised machine learning (Ratner, et al, ArXiv, 2017).
To represent time series information, we framed each patient's timeline as a network (or “graph”) of events. The primary GNN model was a heterogenous graph transformer classifier with two node types: complete blood count (CBC) data and pathology report data (Hu, et al, ArXiv, 2020). Data from the same week were assumed to be from the same timeframe and connected with bidirectional edges. Data separated by longer time periods were connected with unidirectional edges of a separate edge type. The independent test dataset consisted of patients whose ELN 2022 classification was available, and to train the model, the remaining data were divided into train/validation splits of 0.9/0.1.
Results:
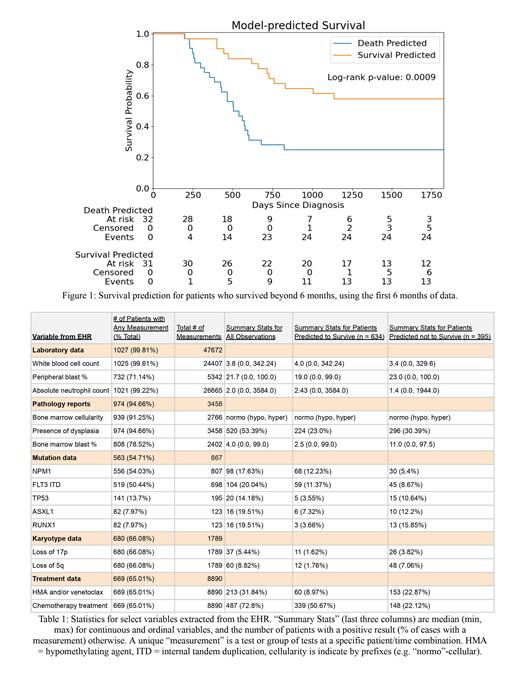
Of the 2,535 patients with survival data, 1,029 met inclusion criteria. Table 1 summarizes the data available in the EHR for each variable, and nearly all patients had CBC and pathology report data. The area under the receiver operating characteristic (AUROC) using the ELN 2022 criteria for predicting survival in the test dataset was 0.79. The AUROC curve for the GNN model was comparable at 0.76, despite not using any variables from the ELN criteria, and the model effectively stratified patients' disease into high- and low-risk in the independent test dataset (hazard ratio [HR] 3.0, log-rank p = 0.0009). Interestingly, despite not having access to mutation or cytogenetic data, the high-risk cases were enriched in known high-risk mutations, like TP53 and RUNX1, and in high-risk chromosomal aberrations, like 5q deletion (Table 1). Although the model predictions correlated with the ELN criteria in some ways, they also stratified the ELN intermediate-risk AML cases into high and low risk (HR 6.1 for model-predicted high risk among ELN intermediate cases, p = 0.07).
Conclusions:
Risk stratification using artificial intelligence and longitudinal data from the EHR performed comparably to the ELN 2022 criteria and has the potential to further stratify the ELN categories. The model performed well despite only using histological features and lab values, which are more readily available and more frequently updated than next-generation sequencing results. In the future, this approach may further improve with a larger sample size and additional variables, such as measurable residual disease and treatment information. Given the heterogeneity and increasing complexity of AML classification, leveraging artificial intelligence to assist with classification will be crucial, and these results are a step towards a future where data are automatically extracted from the EHR and used for continuously updated risk stratification.
Disclosures
Sinha:Verily Life Sciences: Ended employment in the past 24 months. Kuo:Genentech: Current Employment. Mannis:Abbvie: Consultancy; Agios: Consultancy; Macrogenics: Honoraria; Astellas: Consultancy; BMS/Celgene: Consultancy; Genentech: Consultancy; Stemline: Consultancy. Majeti:MyeloGene: Current equity holder in private company; Pheast Therapeutics: Current equity holder in private company; 858 Therapeutics: Membership on an entity's Board of Directors or advisory committees; Orbital Therapeutics: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; kodikaz Therapeutic Solutions: Membership on an entity's Board of Directors or advisory committees. Chen:Google, Inc.: Research Funding. Zhang:Servier: Consultancy; Bristol Myers Squibb: Research Funding; Rigel: Consultancy; Stanford University: Current Employment; Abbvie: Consultancy.