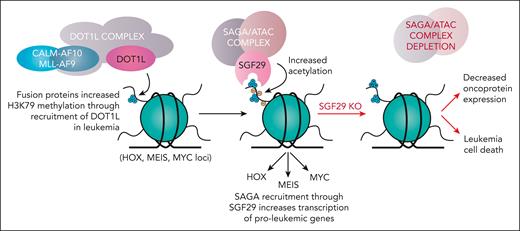
In this issue of Blood, Barbosa et al1 discover that the chromatin reader protein SGF29 is a novel dependency for leukemias with upregulation of the MEIS1/HOX pathway. Chromosomal translocations involving MLL (the mixed lineage leukemia gene) as well as CALM (the clathrin assembly lymphoid myeloid gene) are enriched in infant/pediatric and therapy-related2 leukemias and are often correlated with worse outcomes. MLL- and CALM-rearranged leukemias are characterized by upregulation of the posterior homeobox A (HOX A) gene cluster through recruitment of epigenetic modifiers,3 the best described being DOT1L, a histone H3 lysine 79 (H3K79) methyltransferase.4 Mechanistic dissection of how MLL fusion oncoproteins drive gene dysregulation and leukemia has resulted in several novel therapeutic for leukemias. For instance, discovery of the requirement of DOT1L and menin for HOXA gene upregulation in MLL-rearranged leukemias has led to clinical development of DOT1L and menin inhibitors (the latter of which appear very promising for the treatment of acute myeloid leukemia [AML]5).
Although expression of posterior HOXA genes causes expansion of hematopoietic stem cells,6 it is also known that HOXA genes require coexpression of the related homeobox protein MEIS1 to drive overt leukemia development.7 Armed with this knowledge, Barbosa et al utilized an innovative approach of performing phenotypic drug and CRISPR screens to identify factors that silence expression of MEIS1 in an AML cell line expressing the CALM-AF10 fusion. Specifically, the authors tagged the endogenous MEIS1 gene with a sequence encoding an in-frame green fluorescent protein (GFP), which enabled them to perform high-throughput screens of genes and proteins that led to silencing of GFP.
The author’s initial drug screen utilized previously known drugs targeting an array of epigenetic modifying enzymes. However, the previously known drugs did not affect MEIS1-GFP expression, which led the authors to use an epigenetic enzyme-targeted CRISPR library to broaden their search. Results from their CRISPR knockdown screen identified genes involved in 6 highly enriched chromatin complexes that appeared to regulate MEIS1 expression, including DOT1L, ENL, and CK2 as well as multiple members of the SAGA, KMT2A, and HBO complexes. These top hits were then evaluated with an orthogonal CRISPR screen in single cells, allowing for simultaneous evaluation of genetic suppression and readout of gene expression. This additional screen identified the gene SGF29 (SAGA-associated factor of 29 kDa), which both downregulated MEIS1 and other oncogene expression while simultaneously increasing myeloid differentiation gene expression signatures. Importantly, when the top hits, including SGF29, were validated across AML cell lines, they limited proliferation of MEIS1/HOXA upregulated forms of AML including CALM- and MLL-rearranged AML cell lines.
SGF29 is a member of multiple chromatin regulatory complexes, including the SAGA (Spt-Ada-Gcn5-acetyltransferase) and ATAC (ADA2A-containing) complexes. SGF29 contains 2 tandem C-terminal Tudor domains, which bind histone H3 lysine 4 trimethyl (H3K4me3), a histone modification enriched at promoters, and recruits the SAGA complex for further H3 acetylation (see figure). As such, SGF29 appears critical for promoting chromatin accessibility at several promoters. Here the authors used mutagenesis to identify that SGF29’s Tudor domains are essential for its proleukemic effects. Excitingly, recent work has suggested that the SGF29 H3K4 recognition pocket may be a viable drug target in MEIS1/HOXA-dependent leukemias,8 further underscoring the importance of the discovery of SGF29 as a genetic dependency in AML here.
SGF29 is required for the survival of MEIS1/HOXA upregulated leukemias. CALM- and MLL-rearranged leukemias bind the H3K79 methyltransferase DOT1L and its partner proteins to increase H3K79 methylation across different target gene loci. SGF29 recognizes H3K4 trimethylation, a mark most enriched at gene promoters, through its Tudor domains and recruits the SAGA and ATAC complexes to increase histone H3 lysine acetylation onto target gene loci. This causes increase in downstream gene expression of pro-leukemic genes (including MEIS1). Knockout (KO) of SGF29 prevents SAGA/ATAC complex accumulation at target loci and the subsequent reduction in leukemogenic gene expression results in death of leukemia cells.
SGF29 is required for the survival of MEIS1/HOXA upregulated leukemias. CALM- and MLL-rearranged leukemias bind the H3K79 methyltransferase DOT1L and its partner proteins to increase H3K79 methylation across different target gene loci. SGF29 recognizes H3K4 trimethylation, a mark most enriched at gene promoters, through its Tudor domains and recruits the SAGA and ATAC complexes to increase histone H3 lysine acetylation onto target gene loci. This causes increase in downstream gene expression of pro-leukemic genes (including MEIS1). Knockout (KO) of SGF29 prevents SAGA/ATAC complex accumulation at target loci and the subsequent reduction in leukemogenic gene expression results in death of leukemia cells.
In toto, the authors have discovered a novel target crucial for leukemogenic upregulation of MEIS1 and HOXA genes commonly driving leukemias with MLL and CALM gene rearrangements. Although the authors have provided compelling DepMap, cell line, and patient xenograft data to suggest that knockout of SGF29 may be selectively toxic to leukemic cells, it would next be important to develop means to more stably genetically delete SGF29 (eg, through the use of a conditional knockout mouse of SGF29) to determine its requirement in normal versus malignant hematopoiesis and the therapeutic index of potential SGF29 targeting. Furthermore, it would be important to delineate if additional SGF29-associated proteins in the SAGA and ATAC complexes are also selectively required in HOXA/MEIS1-driven AMLs. Given that each of the SAGA and ATAC complexes contain further potentially druggable enzymes, such an effort may nominate additional novel therapeutic targets for HOX/MEIS1-dependent AMLs. Altogether, this work makes targeting SGF29 through its Tudor domain an attractive target for further development.
Conflict-of-interest disclosure: The authors declare no competing financial interests.