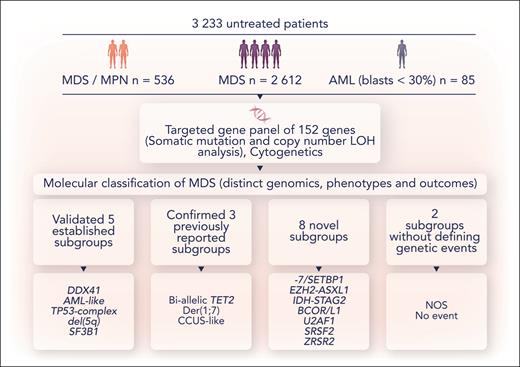
In this issue of Blood, Bernard et al describe their use of a 152-gene targeted next-generation sequencing panel in conjunction with cytogenetics in a large cohort of patients with myelodysplastic syndromes (MDSs) (n = 2612) and related myeloid neoplasms (oligoblastic acute myeloid leukemia [AML], n = 85; and myelodysplastic/myeloproliferative neoplasm [MDS/MPN], n = 536) to derive a novel genomic classification with shared phenotype and outcome associations (see figure).1
A schematic overview of the methodology and conclusions from Bernard et al. NOS, molecularly not otherwise specified; No event, absence of any recurrent drivers evaluated in this study. Professional illustration by Somersault18:24.
A schematic overview of the methodology and conclusions from Bernard et al. NOS, molecularly not otherwise specified; No event, absence of any recurrent drivers evaluated in this study. Professional illustration by Somersault18:24.
The MDSs are currently classified using the 2 major classification systems: the World Health Organization and the International Consensus Classification.2,3 Although there is recognition of some distinct genomic entities in their most recent iterations, namely del(5q), TP53-mutated, and SF3B1-mutated MDS, these account for less than one-third of cases and the remainder of MDS cases are defined by the blast count. In addition, chronic myelomonocytic leukemia and other MDS/MPN entities are defined almost exclusively by phenotype rather than genotype.2-4 The purpose of these classification systems is to define biologically unique entities, but they currently do not reflect the genomic heterogeneity in MDS and the genomic overlap with MDS/MPN entities. Importantly, classification systems are distinct from prognostic models (eg, the Molecular International Prognostic Scoring System for Myelodysplastic Syndromes [IPSS-M]5) whose sole utility is to predict outcome and therefore assist with risk-adapted therapeutic decisions using a limited arsenal of noncurative therapies outside of allogeneic bone marrow transplantation.
Bernard et al used the presence or absence of genetic aberrations (mutations, copy number loss of heterozygosity and aneuploidy), comutations, and the a priori use of TP53 allelic state or IDH2 R140 vs R172 status as inputs for unsupervised clustering to examine complex gene-gene interactions to identify distinct genomic groups (see figure). The output from clustering was then used to derive a hierarchical classification system from 21 genes, 6 cytogenetic events, and 2 allelic states (TET2 and TP53). The authors found 16 genomically defined subgroups that classified 86% of patients with distinct genotype-phenotype associations and clinical outcome, confirming 5 established entities and 3 previously reported entities in addition to discovering 8 new subgroups. Two residual groups were defined either by the absence of group defining or any events. All 18 genomic entities were identified in patients with MDS and chronic myelomonocytic leukemia and many were present in other subtypes of MDS/MPN, albeit at differing frequencies.
Within novel groups, several clinically relevant findings were made. In the −7/SETBP1 group, GATA2 variants were prevalent, with variant allelic frequencies suggestive of a germ line origin in 4%. Although the targeted panel did not include SAMD9 or SAMD9L and germ line tissue was not available, the findings highlight the need to further dissect this genomic group likely to be enriched for patients with a germ line predisposition. The EZH2-ASXL1 group had a high genomic complexity with most patients harboring at least 5 mutations. Both −7/SETBP1 and EZH2-ASXL1 subgroups had poor survival outcomes that were independent of blast count. This was also true for DDX41 and AML-like groups, demonstrating the prognostic power of genomics. IDH-STAG and BCOR/L1 groups were associated with elevated blasts and a high incidence of leukemic transformation with genomic landscapes reminiscent of secondary AML. Overall, these findings do not favor the use of blast percentage to define diagnostic boundaries in these groups.
Comparison between de novo cases and a cohort of therapy-related and postaplastic anemia/paroxysmal nocturnal hemoglobinuria MDS cases demonstrated the overarching influence of genetic lesions in determining phenotype and survival. The unclassifiable groups were typified by mild phenotypes and favorable outcome, where males in the subgroup with no identifiable genetic lesions were enriched for UBA1 mutations along with a phenotype of vacuoles, E1 enzyme, X-linked, autoinflammatory somatic syndrome (VEXAS). Whole-genome sequencing of a subset of patients with elevated blasts and no identifiable driver by panel sequencing yielded putative drivers, including frameshift variants in ATR, FANCC, INPPL1, and RTEL1 with a suspected germ line origin. The impact of genomic modulators of phenotype was elegantly illustrated in patients with biallelic TET2 mutations (13%). Comutation with spliceosome genes was prevalent in 80% of these patients, with modulation of phenotype by ASXL1 and RAS mutations driving monocytosis and JAK2 driving thrombocytosis.
Bearing in mind that this is not a consensus document for a new MDS classification but a proposal for further consideration, Bernard et al provide a foundation for a more refined MDS classification system that may better reflect the underlying biology of these neoplasms. It serves as a steppingstone for future research to focus on understanding the functional consequences of these genomic events and identifying biological differences or similarities between the proposed subgroups. Such findings could be leveraged to identify specific vulnerabilities within certain subgroups to aid drug discovery. However, it is not meant to replace the IPSS-M, which has retained superior predictive performance compared with a model based on these molecular groups.
There are some important limitations to the analysis. Bernard et al used targeted sequencing of 152 genes from peripheral blood or bone marrow with no germ line tissue correlates. Indeed, using their gene panel they would have missed UBTF tandem duplications, an important variant in this context.6 MDS is a hematopoietic stem cell disorder wherein mutant cells contribute to most mature cells in circulation despite perturbations in their capacity to differentiate.7 A genome-wide approach to detect minor variants may have altered this classification system to some degree, but somatic variants are unlikely to account for all the biological heterogeneity in MDS and related myeloid neoplasms, where epigenetic mechanisms likely add to clonal and phenotypic differences that contribute to clinical presentation, disease progression, and treatment response.8,9 In addition, age-related biological differences may exist within the bone marrow microenvironment that alter the trajectory of mutant hematopoietic stem cell proliferation and differentiation.10 With the advent of single-cell multiomics to interrogate cell-type–specific transcriptomic and epigenetic differences in mutant cells, our knowledge of MDS will continue to evolve.
Taken together, this comprehensive analysis has produced a wealth of new information and a framework for a more nuanced classification of MDS.
Conflict-of-interest disclosure: J.E.P. has served on advisory boards and received honoraria or clinical trial funding from Celgene/BMS and AbbVie. L.V. declares no competing financial interests.