HLA-DP permissive mismatches can be assigned a direction according to their immunopeptidome divergence across core and noncore subsets. Noncore permissive graft-versus-host mismatches show significantly reduced risks of relapse without increased nonrelapse mortality compared with allele-matched pairs.
TO THE EDITOR:
The biological basis of permissiveness of HLA-DPB1 mismatches in hematopoietic cell transplantation (HCT)1,2 is related to the nature of the peptide repertoires (ie, immunopeptidomes) presented by these molecules.3 We recently showed that frequent permissive mismatches can be functionally and clinically stratified according to their immunopeptidome divergence into core vs noncore subgroups.4 Permissive mismatches involving the high-frequency core T-cell epitope group 3 (TCE3) alleles (ie, DPB1*02:01, 04:01, and 04:02) associate with lower in vitro T-cell alloresponses and drive the associations with reduced risk of acute graft-versus-host disease (aGVHD) and transplant-related mortality compared with nonpermissive mismatches.4 Interestingly, the stratification of permissive mismatches in these subgroups allows for the introduction of a directionality vector in their assessment. Based on previous evidence for the relevance of HLA mismatches directionality in HCT5,6 and the immunopeptidomic divergence between core and noncore alleles7,8 relevant for alloreactive donor T cells,4,9 we hypothesized that permissive host-versus-graft (HVG) vs graft-versus-host (GVH) HLA-DPB1 mismatches may have different associations with HCT outcome.
To test this novel hypothesis, we applied a refined definition of the permissive subsets in a large contemporary cohort (N = 8456) of well-matched (ie, 10/10) unrelated donor (UD) transplants collected in the EBMT Registry. Patients received transplantation between 2005 to 2020 and had a diagnosis of acute leukemia or myelodysplastic neoplasms (MDS). Detailed information on the patient cohort and clinical data are provided in supplemental Table 1, available on the Blood website. A description of HLA typing and HLA-DPB1 matching, the stratification and directionality of permissive mismatches, and their association with clinical outcomes can be found in the supplemental Materials. We scored all patient-donor pairs as HLA-DPB1 matched (n = 2405) or permissively (n = 3443) or nonpermissively mismatched (n = 2608). Among permissively mismatched pairs, those involving exclusively TCE3 mismatched alleles (n = 3189) were further stratified into core (n = 1642) and noncore nMs (n = 1547; supplemental Tables 2-3; supplemental Figure 1). The directionality of permissive mismatches was defined by the presence (GVH) or absence (HVG) of a mismatched noncore TCE3 allele in the patient. The clinical data were collected and stored by the EBMT Registry. Transplant centers reporting to the registry commit to obtain informed consent in agreement with the principles of the Declaration of Helsinki and according to the local regulations. The study was approved by the Cellular Therapy and Immunobiology Working Party of the EBMT.
Congruent with the established role of the standard TCE model in UD-HCT,10-12 nonpermissively mismatched pairs were associated with significantly increased risks of nonrelapse mortality (NRM; adjusted hazard ratio [HR], 1.26; 99% confidence interval [CI], 1.05-1.50; P = .001) compared with allele matches, which was not observed for permissive mismatches (HR, 1.12; 99% CI, 0.94-1.33; P = .09). Both permissive (HR, 1.23; 99% CI, 1.07-1.41; P = .0001) and nonpermissive mismatches (HR, 1.40; 99% CI, 1.21-1.61; P < .0001) were associated with increased risks of grades 2 to 4 aGVHD and lower relapse (permissive mismatches [HR, 0.87; 99% CI, 0.77-0.99; P = .007]; nonpermissive mismatches [HR, 0.77; 99% CI, 0.67-0.88; P < .0001]) compared with allele matches. Nonpermissive mismatches also significantly increased the risk of severe (ie, grades 3-4) aGVHD (HR, 1.48; 99% CI, 1.16-1.90; P < .0001), whereas this was not statistically significant for permissive mismatches (HR, 1.25; 99% CI, 0.99-1.59; P = .014; Table 1; supplemental Figure 2). No significant associations were observed for other end points (supplemental Table 4).
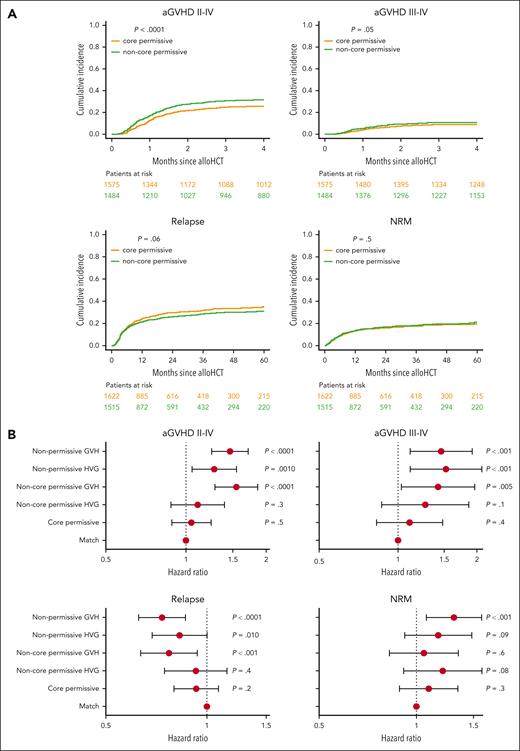
When permissive pairs were stratified into core and noncore mismatches, only noncore permissive pairs showed significantly increased risk of aGVHD (HR, 1.36; 99% CI, 1.16-1.60; P < .0001) and reduced risk of relapse (HR, 0.83; 99% CI, 0.71-0.97; P = .003), whereas for core permissives, neither aGVHD (HR, 1.05; 99% CI, 0.88-1.24; P = .5) nor relapse (HR, 0.93; 99% CI, 0.80-1.08; P = .2) risks differed significantly from allele-matched pairs (Table 1; supplemental Figure 3). Similarly, only noncore mismatches (HR, 1.36; 99% CI, 1.03-1.80; P = .005) but not core permissive mismatches (HR, 1.11; 99% CI, 0.83-1.48; P = .4) significantly increased the risks of severe aGVHD compared with allele-matched pairs. Conversely, neither noncore (HR, 1.11; 99% CI, 0.90-1.37; P = .2) nor core permissives (HR, 1.09; 99% CI, 0.89-1.34; P = .3) associated with significantly higher risks of NRM (Table 1; Figure 1A). As for the standard TCE model, no statistically significant associations were observed for other end points in the core/noncore model (supplemental Table 4).
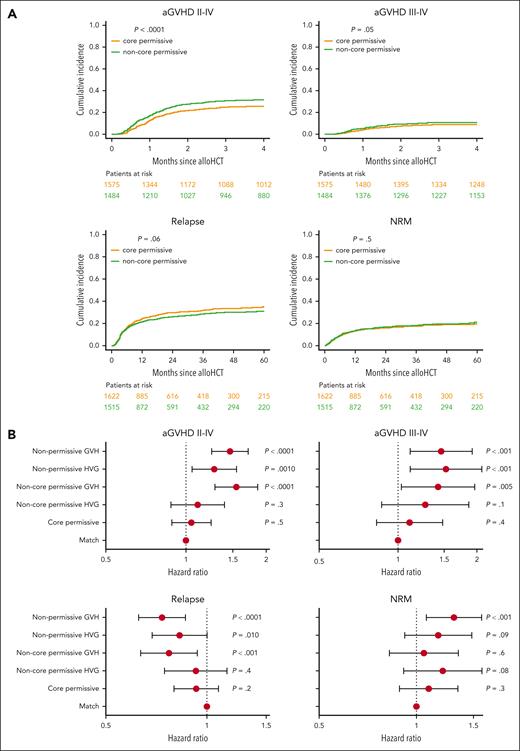
Directional permissive HLA-DPB1 mismatch subsets show differential associations with HCT outcome. (A) Cumulative incidences of grades 2 to 4 and 3 to 4 aGVHD, disease relapse, and NRM are plotted for HLA-DPB1 core permissive (yellow line) and noncore permissive (green line) mismatched pairs. P values indicated correspond to the univariable test of differential cumulative incidences between the 2 patient groups. Patients at risk at different time points are indicated for each group below each plot using the respective color coding. (B) Forest plots show the HR point estimates and 99% CI for grades 2 to 4 and 3 to 4 aGVHD, disease relapse, and NRM, and their associated P values from adjusted multivariable models. HLA-DPB1 permissive and nonpermissive mismatches were scored according to the core/noncore TCE model, in the GVH and HVG directions. Allele-matched patient-donor pairs are taken as reference. alloHCT, allogeneic HCT.
Directional permissive HLA-DPB1 mismatch subsets show differential associations with HCT outcome. (A) Cumulative incidences of grades 2 to 4 and 3 to 4 aGVHD, disease relapse, and NRM are plotted for HLA-DPB1 core permissive (yellow line) and noncore permissive (green line) mismatched pairs. P values indicated correspond to the univariable test of differential cumulative incidences between the 2 patient groups. Patients at risk at different time points are indicated for each group below each plot using the respective color coding. (B) Forest plots show the HR point estimates and 99% CI for grades 2 to 4 and 3 to 4 aGVHD, disease relapse, and NRM, and their associated P values from adjusted multivariable models. HLA-DPB1 permissive and nonpermissive mismatches were scored according to the core/noncore TCE model, in the GVH and HVG directions. Allele-matched patient-donor pairs are taken as reference. alloHCT, allogeneic HCT.
We evaluated the outcomes of directional permissive mismatches along with those of directional nonpermissive subsets.13 Although both HVG and GVH nonpermissives significantly increased the risks of grades 2 to 4 (HVG [HR, 1.28; 99% CI, 1.05-1.55; P = .001] and GVH [HR, 1.46; 99% CI, 1.25-1.71; P < .0001]) and 3 to 4 aGVHD (HVG [HR, 1.52; 99% CI, 1.11-2.09; P < .001] and GVH [HR, 1.46; 99% CI, 1.11-1.92; P < .001]), only nonpermissive GVH mismatches associated with significantly reduced relapse (HVG [HR, 0.83; 99% CI, 0.69-1.00; P = .010] and GVH [HR, 0.73; 99% CI, 0.63-0.83; P < .0001]) and increased NRM (HVG [HR, 1.17; 99% CI, 0.92-1.48; P = .09] and GVH [HR, 1.31; 99% CI, 1.07-1.59; P < .001]) compared with allele-matched pairs (Table 1; Figure 1B).
Among permissive subsets, only noncore permissive GVH mismatches were associated with a significant reduction in relapse risk (HVG [HR, 0.93; 99% CI, 0.75-1.15; P = .4] and GVH [HR, 0.77; 99% CI, 0.63-0.94; P < .001]) and significantly increased risks of aGVHD grades 2 to 4 (HVG [HR, 1.11; 99% CI, 0.88-1.39; P = .3] GVH [HR, 1.55; 99% CI, 1.28-1.86; P < .0001]) and 3 to 4 (HVG [HR, 1.27; 99% CI, 0.87-1.86; P = .1] and GVH [HR, 1.42; 99% CI, 1.03-1.96; P = .005]) compared with allele matches (Table 1; Figure 1B). Importantly, none of the permissive subsets associated with significantly increased risks of NRM (core permissive [HR, 1.09; 99% CI, 0.89-1.34; P = .3], noncore permissive HVG [HR, 1.21; 99% CI, 0.91-1.59; P = .08], and noncore permissive GVH [HR, 1.05; 99% CI, 0.82-1.35; P = .6]; Table 1). Core permissive mismatches showed the lowest probabilities of aGVHD (grades 2-4, 25.8% [95% CI, 23.6%-28.0%]; grades 3-4, 9.0% [95% CI, 7.6%-10.5%] at 4 months after transplant), whereas noncore permissive GVH mismatches had the lowest combination of relapse (28.6% [95% CI, 25.3%-31.9%]) and NRM (20.8% [95% CI, 17.7%-23.9%]) probabilities at 60 months after transplant. This resulted in advantageous relapse-free survival (RFS, 50.6% [95% CI, 46.7%-54.4%] at 60 months) for noncore permissive GVH mismatches compared with all other groups (supplemental Figure 4).
The novel concept of directionality of permissive HLA-DPB1 mismatches explored in this study revealed differential associations with outcomes according to the presence or absence of noncore alleles in the patients. Similarly, our recent study examining peptide-binding motif matching for class I also supports a relevant role for mismatches directionality in clinically relevant alloresponses.6 Our results for HLA-DPB1 suggest that the noncore permissive GVH mismatches could drive more effective graft-versus-leukemia responses than mismatches at other loci, resulting in lower relapse risk in this subgroup without significant NRM. This finding is in accordance with the concept of immunopeptidome divergence modulating alloreactive responses responsible for the GVH/graft-versus-leukemia balance after allogeneic HCT.3,4,7,8
A previous study14 identified a subset of TCE-permissive, high-expression mismatches that were associated with better RFS. Because the majority of frequent noncore TCE3 alleles are linked to the high-expression polymorphism,15 there is a partial overlap between the TCE-permissive, high-expression and the noncore permissively mismatched pairs. However, different to the expression model,11,16 which is only applicable to a subset of transplants, the stratification of permissive mismatches into core and noncore subsets can be applied to all pairs, providing useful clinical information for all patients evaluated while integrating the overlapping information from other models.11,14,16-19
One relevant aspect is that this study did not include patients treated with post transplantation cyclophosphamide (PTCy)-based GVHD prophylaxis.20 Whether the associations with transplant outcome for the standard and core/noncore TCE models are also observed under PTCy and other novel GVHD agents should be investigated once sufficient numbers of evaluable patients are available.
In conclusion, we have shown for the first time that directional, noncore permissive GVH mismatches confer reduced risks of malignant disease relapse, and also independently validated our previous observation that core permissive mismatches at HLA-DPB1 associate with reduced GVHD after UD-HCT. This suggests the possibility of tailored UD selection according to the main clinical priority in patients with hematologic malignancies (ie, GVHD risk minimization vs relapse risk reduction) and potentially also for patients treated for nonmalignant diseases. Further studies to confirm the potential clinical advantages of directional core vs noncore permissive mismatches, in particular in the context of novel strategies for GVHD prevention including PTCy20 and of mismatches at other HLA loci,21 are warranted.
Acknowledgments
The authors thank the staff at the participating transplant centers collecting and contributing clinical data for this study.
This work was supported by grants from the Deutsche Knochenmarkspenderdatei (DKMS-SLS-JHRG-2021-02 [E.A.-B.]) and the Deutsche José Carreras Leukämie Stiftung (DJCLS 20R/2019), the Werner Jackstädt Stiftung, the Joseph Senker Stiftung, and the Deutsche Forschungsgemeinschaft (DFG FL 843/1-1) (K.F.).
Authorship
Contribution: E.A.-B. and K.F. designed the study; T.G.-D., M.R., U.S., N.K., I.Y.-A., A.H., C.C., E.D., C.E.B., E.F., and E.T. contributed clinical data; L.L.J.v.d.B., L.C.d.W., and E.A.-B. performed statistical analyses; E.A.-B., K.F., L.L.J.v.d.B., L.C.d.W., A.R., and J.D.H. analyzed and interpreted data; E.A.-B. drafted the manuscript; and all authors participated in manuscript writing and review and provided final approval of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Esteban Arrieta-Bolaños, Institute for Experimental Cellular Therapy, University Hospital Essen, Hufelandstr 55, 45122 Essen, Germany; email: esteban.arrieta-bolanos@uk-essen.de.
References
Author notes
F.M. and A.R. are the secretary and chair of the CTIWP of the EBMT, respectively.
A.R. and K.F. contributed equally to this study.
Data are available on request from the EBMT Cellular Therapy and Immunobiology Working Party Chair (ctiwp@ebmt.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.